Abstract
The human chromosome 22q11.2 region is susceptible to rearrangements during meiosis leading to velo-cardio-facial/DiGeorge/22q11.2 deletion syndrome (22q11DS) characterized by conotruncal heart defects (CTDs) and other congenital anomalies. The majority of individuals have a 3 Mb deletion whose proximal region contains the presumed disease-associated gene TBX1 (T-box 1). Although a small subset have proximal nested deletions including TBX1, individuals with distal deletions that exclude TBX1 have also been identified. The deletions are flanked by low-copy repeats (LCR22A, B, C, D). We describe cardiac phenotypes in 25 individuals with atypical distal nested deletions within the 3 Mb region that do not include TBX1 including 20 with LCR22B to LCR22D deletions and 5 with nested LCR22C to LCR22D deletions. Together with previous reports, 12 of 37 (32%) with LCR22B–D deletions and 5 of 34 (15%) individuals with LCR22C–D deletions had CTDs including tetralogy of Fallot. In the absence of TBX1, we hypothesized that CRKL (Crk-like), mapping to the LCR22C–D region, might contribute to the cardiac phenotype in these individuals. We created an allelic series in mice of Crkl, including a hypomorphic allele, to test for gene expression effects on phenotype. We found that the spectrum of heart defects depends on Crkl expression, occurring with analogous malformations to that in human individuals, suggesting that haploinsufficiency of CRKL could be responsible for the etiology of CTDs in individuals with nested distal deletions and might act as a genetic modifier of individuals with the typical 3 Mb deletion.
Introduction
Congenital heart disease (CHD) is the most common type of birth defect in humans. It occurs in nearly 1% of all live births and is considered the major cause of infant mortality.1 Many individuals require surgical intervention during the first year of life for survival. Overall, the quality of life of individuals with CHD is often compromised. Despite its clinical relevance, the genetic cause of most CHD remains unknown. The majority is of sporadic occurrence perhaps caused by a combination of genetic and environmental risk factors.2 Studies of rare genetic CHD syndromes might help pinpoint genes whose contribution to disease risk and mechanism can be better understood in animal models.
Velo-cardio-facial syndrome (MIM 192430) or DiGeorge syndrome (MIM 188400), also known as 22q11.2 deletion syndrome (22q11DS), is a congenital malformation disorder occurring in 1/4,000 live births.3 It is characterized by extensive phenotypic variability that has been the subject of intensive research. Approximately 70% of affected individuals have a conotruncal type of heart defect (CTD; e.g., tetralogy of Fallot [TOF], interrupted aortic arch type B [IAAB], persistent truncus arteriosus [PTA]) and/or aortic arch anomalies (AAAs; e.g., right-sided aortic arch). More than 90% of individuals have a 3 Mb deletion. The deletion is usually de novo in occurrence and is caused by meiotic non-allelic homologous recombination events between flanking low-copy repeats termed LCR22. There are several LCR22s in the 22q11.2 region;4,5 the most relevant are LCR22A, LCR22B, LCR22C, and LCR22D (Figure 1). The typical 3 Mb deletion is flanked by LCR22A and LCR22D (LCR22A–D). More than 45 known genes map to the 3 Mb interval and are dispersed throughout the region. Approximately 10% of individuals have a proximal nested 1.5 Mb deletion between LCR22A and LCR22B, and 5% have a 2.0 Mb deletion between LCR22A and LCR22C.6,7 The 1.5 Mb LCR22A–B region has been considered the critical region for the syndrome.8,9 TBX1 (T-box 1; Figure 1) (MIM 602054), mapping to the critical LCR22A–B interval, is the strongest candidate gene for CTDs as determined by studies of mouse models.10,11 Furthermore, mutations in TBX1 were discovered in a handful of non-deleted human individuals with a subset of clinical features of 22q11DS.12,13
Figure 1.
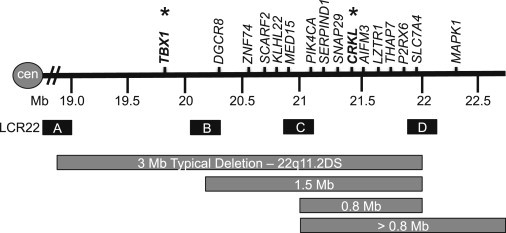
The 22q11.2 Region and LCR22s
Schematic representation of the 22q11.2 region showing the different sizes of deletions. Low-copy repeats on chromosome 22q11.2 (LCR22) are represented by letters A, B, C, D (sizes not to scale). A subset of the genes in the 22q11.2 region is shown.
Studies of mouse models also implicated another gene, the cytoplasmic adaptor CRKL (v-crk avian sarcoma virus CT10 oncogene homolog-like) (MIM 602007), in the etiology of 22q11DS.14 However, CRKL maps to the LCR22C–D interval and is thus outside the presumed critical region (Figure 1). Interestingly, over the past few years individuals with distal LCR22B–D and nested LCR22C–D deletions that do not include TBX1 have been reported.6,7,15,16 There are 11 coding genes (10 known genes and 1 predicted gene) in the LCR22C–D region including PI4KA (phosphatidylinositol 4-kinase, catalytic, alpha [MIM 600286]), SERPIND1 (serpin peptidase inhibitor, clade D [heparin cofactor], member 1 [MIM 142360]), SNAP29 (synaptosomal-associated protein, 29 kDa [MIM 604202]), CRKL, LOC1019288, AIFM3 (apoptosis-inducing factor, mitochondrion-associated, 3), LZTR1 (leucine-zipper-like transcription regulator 1 [MIM 600574]), THAP7 (THAP domain containing 7 [MIM 609518]), P2RX6 (purinergic receptor P2X, ligand-gated ion channel, 6 [MIM 608077]), SLC7A4 (solute carrier family 7, member 4 [MIM 603752]), and LRRC74B (leucine rich repeat containing 74B). Based upon previous mouse genetic studies, CRKL is the strongest candidate gene for the cardiac phenotype in the region and thus worthy of further studies in mouse models.14,17,18
Although Tbx1 has been extensively studied in mouse models, Crkl has been given less attention with regard to 22q11DS. Crkl belongs to the Crk family of cytoplasmic protein adaptors that includes itself19 and Crk (v-crk avian sarcoma virus CT10 oncogene homolog) (CRKI, CRKII, and CRKIII, three spliced variants [MIM 164762]).20,21 Both Crk and Crkl are evolutionally conserved from nematodes to vertebrates and encode ubiquitously expressed proteins that play essential roles in signal transduction, such as MAPK signaling. Of note, CRK maps to human chromosome 17, not 22q11.2. Both CRK and CRKL mediate tyrosine kinase signaling to regulate cell proliferation, differentiation, adhesion, and migration.22 They are also recognized as key components in cancer metastasis and invasion.23,24 Despite their similar sequence, they have independent roles during mammalian development. There have been a few important mouse studies of Crk and Crkl during embryogenesis. In particular, loss of function of either gene results in lethality due to the presence of embryonic malformations including CHD. Crk-null mice have vascular smooth muscle cell defects causing dilatation and rupture of blood vessels.25 They also have thin myocardial walls with normal-appearing conotruncal and valvular structures.25 Crkl homozygous null mutant embryos, on the other hand, have CTDs similar to that seen in 22q11DS-affected individuals.14 Because of the presence of similar CTD malformations in Tbx1- and Crkl-null mutant embryos, and because both genes are hemizygous in most individuals with 22q11DS, heterozygous intercrosses were performed to uncover a possible genetic interaction. Analysis of double heterozygous embryos revealed that Crkl genetically interacts with Tbx1 for cardiovascular development.17 Thus, Crkl might act in the Tbx1 genetic pathway, possibly serving as a genetic modifier in humans with the 3 Mb 22q11.2 deletion.
Given these observations, we decided to reexamine the role that CRKL might play in CHD. We evaluated 25 individuals with a nested atypical LCR22B–D (n = 20) or LCR22C–D (n = 5) deletion that is hemizygously deleted for CRKL and have CTDs but do not have a deletion of TBX1. When taken together with previous reports, there are now 71 subjects described with LCR22B–C or LCR22C–D deletions. Further, we were able to recapitulate the full spectrum of Crkl dosage effects on cardiac outflow tract (OFT) development by creating and evaluating a Crkl allelic series in mice. These data implicate CRKL as an important human disease gene for CTDs.
Material and Methods
Human Study Subjects
A total of 1,232 individuals with 22q11DS were evaluated at The Children’s Hospital of Philadelphia from 1992 to 2014. The individuals had deletion sizing performed by a variety of molecular methods that evolved as technology advanced. Clinical samples, comprising 62% of the cohort, were evaluated by fluorescence in situ hybridization (FISH) and more recently by array comparative genomic hybridization (aCGH), genome-wide SNP microarray, or MLPA. Research samples were evaluated by FISH, or more recently by multiplex-ligation-dependent probe amplification assays (MLPA; MRC Holland; P023 or P250 SALSA). Echocardiography reports were reviewed for each subject. All investigations were conducted under an IRB-approved protocol at The Children’s Hospital of Philadelphia and Albert Einstein College of Medicine (CCI 1999-201). Informed consent was obtained for all participants.
Generation of Crkl Alleles
We created a Crkl-floxed and Crkl-null allele in mice as shown in Figure S1. To generate a Crkl floxed allele, we first inserted a single loxP site upstream of the first exon of Crkl (Figure S1; yellow). We inserted a Neomycin cassette flanked by two loxP sites downstream of exon 1 (Figure S1; orange and green). There were a total of three loxP sites in the Crkl locus, and the mice were termed Crkl-NEO/+. To generate a null allele, we crossed floxed mice with CMV-Cre mice and screened resulting animals by PCR. The mouse lines used have been generated for the purpose of this study and they are Crkl-NEO/+ and Crkl+/− or Crkl-HET. We backcrossed each line or intercrossed them in order to obtain all the possible allelic combinations: Crkl+/+ or Crkl-WT, Crkl-HET, Crkl-NEO/+, Crkl-NEO/NEO, Crkl-NEO/−, and Crkl−/− or Crkl-KO. All mice have been maintained congenic in the Swiss Webster background. PCR strategies for mouse and embryos genotyping are available upon request.
Quantitative RT-PCR
To obtain enough total RNA and minimize the variability of gene expression in individual embryos, each RNA sample contained microdissected hearts from three embryos of the same genotype at E10.5. Three biological replicates were performed per genotype. The tissue was isolated and immediately frozen, and samples were homogenized in Buffer RLT (QIAGEN). Total RNA was isolated with the RNeasy Micro Kit (QIAGEN) according to the manufacturer’s protocol. Quality and quantity of total RNA was determined with an Agilent 2100 Bioanalyzer (Agilent) and a ND-1000 Spectrophotometer (NanoDrop), respectively. Single-stranded cDNA targets were amplified from 100 ng starting total RNA with the Ovation RNA Amplification System V2 and FL-Ovation cDNA Biotin Module V2 (NuGEN). The mRNA levels were measured by TaqMan Gene Expression Assays (Applied Biosystems) for each gene and were carried out in triplicate with 18S (RNA, 18S ribosomal 1), Actb (Actin, beta), and B2m (Beta-2-microglobulin) genes as normalization controls. TaqMan probes and primer sets were obtained from the Applied Biosystems Gene Expression Assay database. Samples were run in standard 96-well plates (20 μl final volume per reaction and each reaction in triplicate) on an ABI 7900HT Q-PCR apparatus. The SDS 2.2 software platform (Applied Biosystems) was used for the computer interface with the ABI 7900HT PCR System to generate normalized data, compare samples, and calculate the relative quantity. Statistical significance of the difference in gene expression was estimated by the two-tailed t test.
Mouse Heart Histology and Phenotypic Analysis
Mouse embryos were isolated in PBS and fixed in 10% neutral buffered formalin (Sigma) overnight. After fixation, the embryos were dehydrated through a graded ethanol series, embedded in paraffin, and sectioned at 5 μm. All histological sections were stained with hematoxylin and eosin by standard protocols. Staining was performed in the Einstein Histopathology Core Facility. A total of 118 hearts at E15.5 were obtained from approximately 50 independent crosses and analyzed morphologically via light microscopy.
Phenotype-Genotype Heatmap
For both human and mouse samples, we created a phenotype matrix indicating the cardiac phenotypes found in each sample. We used the heatmap R package to create the phenotype-genotype correlation shown as heatmaps. Clustering distances for x and y axes were calculated by Euclidean distance measures.
Results
Congenital Heart Defects in Individuals with the Nested LCR22B–D Deletion
The LCR22B–D region is 0.75 Mb in size, excluding the flanking LCR22s (chr22: 20,726,074–21,472,008; hg19) or 1.6 Mb including the LCR22s (chr22: 20,274,541–21,921,290). We evaluated 20 newly ascertained, unrelated persons with the 22q11.2, LCR22B–D deletion (Figure 1) with known heart phenotype information based upon echocardiography reports (Table S1). Overall, 8 of the 20 individuals (40%) had cardiac malformations of which half were conotruncal type defects (CTDs). We observed tetralogy of Fallot (TOF; n = 2), persistent truncus arteriosus (PTA; n = 1), interrupted aortic arch type B (IAA type B; n = 1), isolated subaortic ventricular septal defects (VSDs; n = 4), and atrial septal defects (ASDs; n = 2). Two individuals had more than one cardiac anomaly, such as VSD and ASD. We then combined our results with 17 previously published cases, of which 10 were unrelated probands and 7 were family members.6,7,15,26 Taken together, 12/37 (32%) individuals with LCR22B–D deletions had CHD (Figure 2A). To better visualize the comparison, we created a heatmap in which we focused on phenotypes with respect to the position of the deletion on chromosome 22q11.2 (Figure 2B). Among the 12 individuals with LCR22B–D deletions that had CHD, the most common defects were TOF (3/12; 25%) and isolated subaortic VSDs (4/12; 33%). Other less common defects in previously reported persons with LCR22B–D deletions included aortic arch anomalies, PTA, and double outlet right ventricle (DORV) (Figure 2B).
Figure 2.
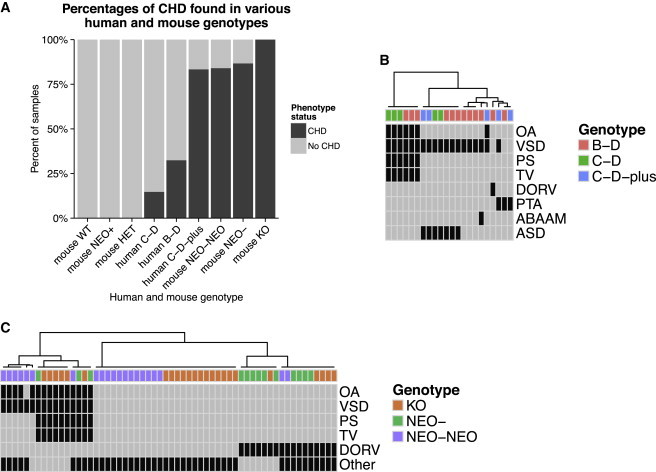
Congenital Heart Defects in Human and Mouse
(A) Percentages of CHD in all available human and mouse genotypes relevant for this study. Genotypes consisted of wild-type, Crkl-WT, hypomorphic (Crkl-NEO/NEO and Crkl-NEO/−), and null (Crkl-KO).
(B and C) Human LCR22B–D deletion is indicated as B-D; LCR22C–D deletion is indicated as C-D, LCR22C to beyond LCR22D is indicated as C-D-plus. Genotype-phenotype heatmaps indicating phenotype status for each subject with CHD are given for human (B) and mouse (C) data.
(B) Human subjects with LCR22B–D or LCR22C–D deletions are shown along the x axis. The various reported cardiac phenotypes are shown on the y axis. The most common phenotype is TOF, which includes an OA with VSD, associated with a hypertrophic right ventricle (thick ventricle; TV) and PS. ABAAM (abnormal branch arch artery morphology) and DORV are seen only in subjects with LCR22B–D deletions.
(C) Mouse samples with different Crkl alleles are shown along the x axis and observed phenotypes are shown along the y axis. Mice with the lowest level of Crkl expression have DORV or TOF. In mouse embryo tissue sections, TOF is defined as the presence of OA, VSD, TV, and PS. Row and column clustering was performed by Euclidean distance measures. Abbreviations are as follows: ASD, atrial septal defect; ABAAM, abnormal branchial (pharyngeal) arch artery morphology; DORV, double outlet right ventricle; PTA, persistent truncus arteriosus; VDS, ventricular septal defect; TV, thick ventricular wall; OA, overriding aorta; and PS, pulmonary stenosis. “Other” represents non-conotruncal cardiac defects.
Cardiac Defects in Individuals with the LCR22C–D Deletion
The LCR22C–D deletion spans 0.4 Mb, excluding the LCR22 sequences (chr22: 21,062,071–21,466,724). Of the five persons ascertained (Table S1) with the LCR22C–D deletion (Figure 1), one of them had TOF and the rest did not have a heart defect. From the literature of 19 unrelated probands and 10 family members, 2 had TOF and 2 had both a VSD and ASD.15–20,26 Thus a total of 5/34 (15%) of individuals with an LCR22C–D deletion had CHD (Figure 2A), including TOF in 3/5 (60%; Figure 2B).
Eight LCR22s span the 22q11.2 region: LCR22A, B, C, D, E, F, G, and H. There are six published cases of atypical deletions flanking LCR22C that extend beyond LCR22D (termed LCR22C–D-plus in this study; reviewed by Rump et al.)26 These deletions include CRKL as well as MAPK1 (mitogen-activated protein kinase 1) (MIM 176948), a CRKL pathway gene (Figure 1). A total of 5/6 (83%) of these individuals had CHD, including a VSD and ASD (Figure 2). Of interest, OA and VSD, comprising two components of TOF (Figure S3), was observed in one previously reported individual with an atypical LCR22C–D-plus deletion that extended beyond LCR22D26 (Figure 2B). Overall, these distal deletions with CRKL further suggest its contribution to CHD in these individuals.
Generation of an Allelic Series with Varying Crkl mRNA Dosage
One important question has been whether the function of CRKL is sensitive to altered gene dosage in humans. Although Crkl-HET mice have no heart defects, Crkl-KO embryos have previously been reported to have VSDs, occasionally with cardiac outflow tract alignment defects such as DORV or an overriding aorta (OA).14,18 In histological sections from late-gestational-stage mouse embryos, TOF is defined as OA, VSD, hypertrophic right ventricle (RV), and pulmonic stenosis (PS). TOF is similarly defined in humans by echocardiography, and in particular requires anterior malalignment of the conal septum with mitral to aortic valve fibrous continuity. An OA and PS are also characteristic, as is hypertrophic RV, occurring as a secondary defect. These working definitions allow for the comparison between the different kinds of data generated from human individuals and mouse embryos.
To try to generate the full spectrum of phenotypes observed in human subjects in a mouse model, we took advantage of the fact that insertion of a Neomycin cassette into introns occasionally interferes with expression of that gene and can produce hypomorphic models.27 We generated a hypomorphic mouse line, Crkl-NEO/NEO (Figure S1), that expresses Crkl mRNA at a lower level than the Crkl-HET mouse line (Figure 3A). To examine the phenotype in the hypomorphic mice, we intercrossed Crkl-NEO/+ mice and found that most Crkl-NEO/NEO mice died before birth (expected frequency, 1:4; observed frequency, 1:30; i.e., three pups with this genotype were found out of 100 pups recovered from at least 10 independent mating pairs). To test whether the Neomycin cassette reduced expression of Crkl mRNA, two Crkl mouse lines, Crkl-HET and Crkl-NEO/+, were crossed to obtain six different allelic combinations. Quantitative RT-PCR was performed as shown in Figure 3B. The presence of the Neomycin cassette reduced expression of Crkl as compared to the normal level (Figure 3B). The heterozygous Crkl-NEO/+ allele had expression levels similar to Crkl-HET values. Embryonic hearts from the Crkl-NEO/NEO genotype had a greatly reduced level of Crkl expression (Figure 3B). Expression levels in Crkl-NEO/NEO and Crkl-NEO/− hearts were not significantly different. Overall, this tells us that the Crkl-NEO allele behaves as a hypomorphic allele.
Figure 3.
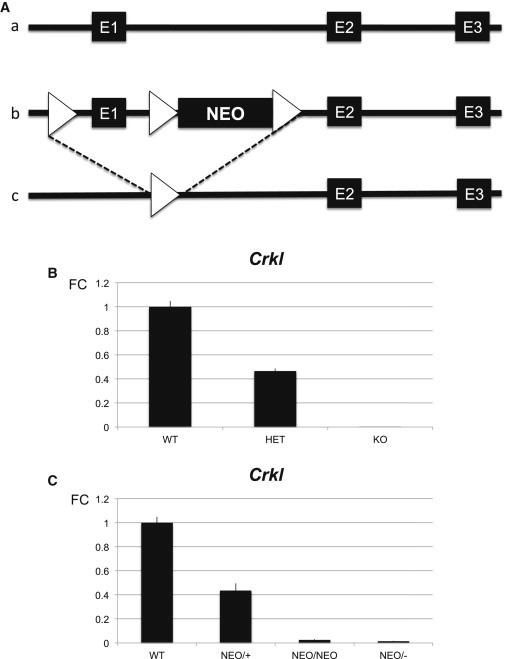
Generation of an Allelic Series with Varying Crkl mRNA Expression
(A) Schematic of the Crkl mouse alleles and cardiac expression. Exons are indicated by large black boxes. An allelic series of Crkl expression was obtained by specific genetic crosses that allowed all possible combination of two of the three alleles described, i.e., a, b, and c. The normal locus (a), the floxed allele (loxP, triangle) containing the Neomycin cassette (NEO) (b), and the recombined allele (c) are shown.
(B and C) The fold change (FC) from quantitative RT-PCR (qRT-PCR) assays to detect Crkl mRNA dosage in mouse hearts at E10.5 isolated from each genotype is indicated. All mice were maintained in a pure Swiss-Webster genetic background. The fold change (FC ± SD) of Crkl mRNA expression in the allelic series with respect to the wild-type were as follows: Crkl-WT (1 ± 0.049), Crkl-HET (0.465 ± 0.02), and Crkl-KO (0.003 ± 0.0001) hearts and Crkl-WT (1 ± 0.049), Crkl-NEO/+ (0.436 ± 0.059), Crkl-NEO/NEO (0.025 ± 0.006), and Crkl-NEO/− (0.013 ± 0.003) alleles are shown. Statistical significance of the difference in gene expression was estimated by the two-tailed t test, p values ≤ 0.01. All cross comparisons between the above-mentioned genotypes were significantly different except for NEO/+ versus HET and NEO/NEO versus NEO/−.
One concern with the Neomycin cassette is that it might modify expression of neighboring genes. This would result in a phenotype unrelated to that of our gene of interest. To test this, we measured the expression levels of eight genes in the LCR22C–D region (Figure 4) by qRT-PCR. We found that the Neomycin cassette did not alter the expression level of any of them (Figure 4), confirming that the Neomycin cassette in this mouse model affected only Crkl expression. Of interest, one of the selected genes, Aifm3, was increased in expression when the dosage of Crkl was decreased. This is unrelated to the presence of the Neomycin cassette, because expression of Aifm3 in Crkl-NEO/NEO and Crkl-KO embryos did not differ (Figure 4).
Figure 4.
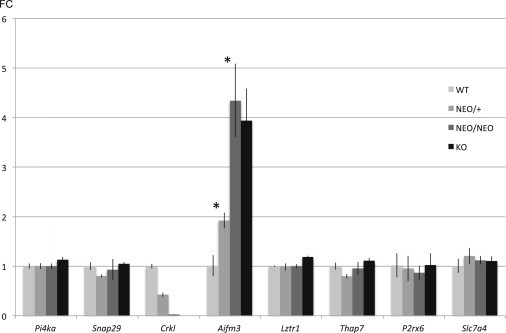
Gene Expression of Crkl Neighboring Genes
Quantitative RT-PCR of genes in the LCR22B–D region flanking Crkl. Similar qRT-PCR assays were performed as in Figures 3B and 3C. Mouse hearts were isolated from Crkl-WT, Crkl-NEO/+, Crkl-NEO/NEO, and Crkl-KO hearts at E10.5. Expression values are shown as fold change and error bars are the standard deviations (FC ± SD). Statistical significance of the difference in gene expression was estimated by the two-tailed t test, p values ≤ 0.01.
Severity of OFT Defects Is Sensitive to Crkl Expression Levels in the Mouse
The Crkl allelic series, in order of decreasing expression, consists of Crkl-WT, Crkl-HET, Crkl-NEO/+, Crkl-NEO/NEO, Crkl-NEO/−, and Crkl-KO genotypes. To delineate the spectrum of heart defects in embryos of the Crkl allelic series, we examined the hearts from 118 embryos at E15.5 (percentages of CHD are shown in Figure 2A; details in Table S2). To better visualize the comparison, we created a heatmap in which we focused on phenotypes with respect to Crkl dosage (Figure 2C). We selected the time point of E15.5 because that is 1 day in embryonic development prior to the death of Crkl-KO embryos.14 The distribution of percentage of different OFT cardiac phenotypes within the groups of similar genotypes is shown in Figure 5. CHD phenotypes were present in 21/25 (84% overall; 32% CTD only) of Crkl-NEO/NEO embryos, 13/15 (87% overall, of which all were CTDs) of Crkl-NEO/− embryos, and 24/24 (100% over all; 46% CTD only) of Crkl-KO embryos. Non-CTD phenotypes included myocardial and valvular defects. The hypomorphic embryos, Crkl-NEO/NEO and Crkl-NEO/−, displayed similar CHD patterns to those found in humans with the distal nested deletions.
Figure 5.
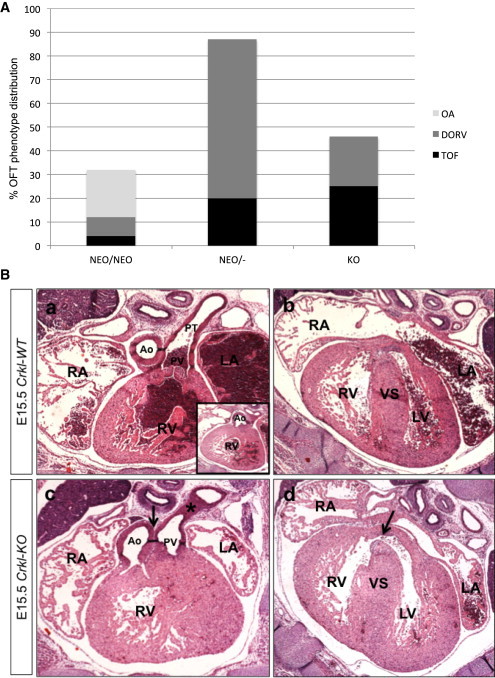
Conotruncal Defects and Crkl Alleles
(A) Categories of cardiac outflow tract defects in Crkl mutant mouse embryos. Percent of embryos with cardiac defects in the three genotypes of Crkl that disrupts cardiac outflow tract development (Crkl-NEO/NEO = 25, Crkl-NEO/− = 15, Crkl-KO = 24). Note that one embryo can have multiple defects (Table S2). Distribution of outflow tract defects TOF, DORV, and OA.
(B) TOF in a Crkl-KO embryo. Transverse histological sections of a Crkl-WT (a, b) and Crkl-KO (c, d) embryonic mouse heart at E15.5 stained with hematoxylin and eosin. Arrow (c) shows misalignment of the Ao and PV. Star (c) shows PS. Arrowhead (d) shows a VSD. Abbreviations are as follows: RA, right atrium; RV, right ventricle; LA, left atrium; LV, left ventricle; Ao, Aorta; PV, pulmonary valve; VS, ventricular septum.
The most common CHD in humans with the LCR22B–D or nested deletions is an isolated subaortic VSD (Figure 2A). In mice, isolated VSDs with normally aligned outflow tracts or associated with a mild alignment defect as OA (Figure S2) occurred with increasing frequency as Crkl expression level increased. In contrast, TOF and DORV, which are severe alignment defects, occurred with increasing frequency as Crkl expression decreased (Figure 5B). Specifically, TOF occurred in 1/25 of Crkl-NEO/NEO embryos (4%), 3/15 in Crkl-NEO/− embryos (20%), and 6/24 of Crkl-KO embryos (25%) (Figure 5B). Similarly, DORV occurred in 2/25 Crkl-NEO/NEO (8%), 10/15 Crkl-NEO/− (67%), and 5/24 Crkl-KO (21%) embryos. In contrast, OA with no other outflow tract defect occurred in 5/25 Crkl-NEO/NEO embryos (20%) but did not occur in Crkl-NEO/− and Crkl-KO embryos. These data suggest that the severity of the cardiac defects in Crkl mutant embryos is sensitive to altered expression. It also indicates that Crkl hypormorph embryos have a similar penetrance and expressivity to that found in human individuals with distal deletions. When taken together, it supports the idea that CRKL is dosage sensitive for function in humans.
Discussion
In this report, we describe individuals with variant chromosome 22q11.2 deletions in the distal half of the 3 Mb region that exclude TBX1, with clinical features of 22q11DS (velo-cardio-facial/DiGeorge syndrome). We refer to these as nested LCR22B–D and LCR22C–D distal deletions. CRKL, mapping to the LCR22C–D region, among other genes is hemizygously deleted. By generating an allelic series, we demonstrated that Crkl is sensitive to reduced gene expression for cardiac OFT development. Overall, our data implicate CRKL as a disease-associated gene for individuals with nested distal 22q11.2 deletions and we suggest that it could act as a modifier of CTDs for those with the typical hemizygous 3 Mb 22q11.2 deletion.
The Mouse Crkl Allelic Series
Among the different alleles we obtained, the hypomorphic Crkl-NEO/NEO and Crkl-NEO/− alleles elicit the full range of different CTDs that occur in individuals with nested distal deletions. Neither the Crkl-HET nor the Crkl-KO embryos are optimal models because they have either no phenotype or additional intracardiac muscle or valve defects, respectively. The cardiac OFT defects between the Crkl-NEO/NEO and Crkl-NEO/− embryos were compared. Of interest, the penetrance and expressivity of particular CTDs were quite different, despite the fact that the expression levels of Crkl in these two allelic combinations were not considered different based on a statistical evaluation. The mice were maintained in the same congenic background, implicating subtle changes in Crkl expression as being responsible for the differences in the phenotype in mutant embryos. This suggests that there is a specific threshold of Crkl expression that when reduced beyond a particular level greatly affects heart development. For example, it is likely that there is a gradient effect for Crkl expression. It might be possible to observe such changes at the protein level or with respect to changes in phosphorylation of downstream effector proteins. Either way, we consider the Crkl hypomorph to be a unique tool to study causes of genetic variability of heart development.
Based upon the frequency of specific malformations present, the hypomorphic alleles might make it possible to understand the molecular etiology of individual cardiac OFT phenotypes. In addition, the Crkl hypomorph mouse models might help to identify genes that act in the same genetic pathway as Crkl. The usual genetic strategy to identify genes that can regulate Crkl expression or act downstream of MAPK signaling involves intercrosses between two different heterozygous mouse alleles. Frequently, double heterozygous mice have a mild phenotype or one with reduced penetrance. The Crkl-NEO/NEO or Crkl-NEO/− mouse model might provide an advantage in such genetic interaction studies because the threshold needed to observe a phenotype is reduced. It also suggests that genetic variations in human CRKL might act as a genetic modifier of CTDs in individuals with the 3 Mb deletion.
Cardiac Genotype to Phenotype Correlations in Individuals with LCR22B–D versus LCR22C–D Deletions
We describe individuals with nested LCR22B–D deletions harboring CTDs as well as those reported in the literature. Such nested deletions are only now being recognized in the clinical lab given the advent of genome-wide molecular testing. One important goal is to determine which gene(s) are responsible for the etiology of the human nested distal deletion phenotype. There are 11 coding genes (10 known and 1 predicted) in the LCR22C–D region: PI4KA, SERPIND1, SNAP29, CRKL, LOC1019288, AIFM3, LZTR1, THAP7, P2RX6, SLC7A4, and LRRC74B (Table S3). One question is whether haploinsufficiency of any of the other genes could contribute to the phenotype in affected individuals. One gene of interest is SNAP29 because it is already known to be associated with a human disease. SNAP29 encodes a synaptic vesicle protein that when mutated causes autosomal-recessive CEDNIK syndrome (cerebral dysgenesis, neuropathy, ichthyosis, keratoderma [MIM 609528]). However, individuals with this disorder do not have CHD. Another gene in the region is SERPIND1, which encodes heparin cofactor II that is important in the blood coagulation cascade28,29 but is not required for heart development. P2RX630,31 and SLC7A432 function in ion channels and are unlikely to contribute to cardiac outflow tract morphogenesis. This leaves six genes remaining, including CRKL. The five other genes are PI4KA, encoding a phosphatidylinositol kinase,33,34 AIFM3, encoding a mitochondrial protein that function in inducing apoptosis,35 LZTR1, encoding a golgi apparatus protein,36 THAP7, encoding a chromatin modifier,37,38 and LRRC74B, encoding a protein of unknown function. Among the LCR22C–D genes besides CRKL, THAP7 seems interesting because of its function as a chromatin modifier needed to repress transcription.38,39 However, it does not appear strongly expressed in the pharyngeal apparatus, which provided progenitor cells to the OFT or heart.40
One interesting gene is AIFM3 because mRNA expression in embryonic hearts was increased in expression when Crkl was inactivated. AIFM3 is related to AIFM1 (apoptosis-inducing factor, mitochrondrian-associated-1 [MIM 300169]) and encodes a ubiquitously expressed mitochondrial protein that induces apoptosis when overexpressed.41 Overexpression of AIFM3 might increase apoptosis, but the possible function of AIFM3 in heart development is not known. Because individuals with LCR22B or LCR22C–D deletions will be hemizygously deleted for AIFM3, the overall expression level might be normalized. Among all the other genes in the region, CRKL is the strongest candidate gene.
Our work suggests that there might be additional genes in the LCR22B–C region that contribute to CHD, because the penetrance is higher in subjects with LCR22B–D than LCR22C–D deletions. The LCR22B–C interval contains four coding genes: ZNF74 (zinc finger protein 74 [MIM 194548]), encoding a protein that might be involved in RNA maturation or transcription,39,42 SCARF2 (scavenger receptor class F, member 2 [MIM 613619]), in which recessive mutations cause Van Den Ende Gupta syndrome (MIM 600920) that is not characterized by CHD,43,44 KLHL22 (kelch-like family member 22), which encodes a protein involved in regulation of chromosome alignment during meiosis,45,46 and MED15 (mediator complex subunit 15 [MIM 607372]), which encodes a protein that is part of the Mediator complex needed for RNA polymerase II transcription.3,7 Among the four genes, MED15 is the most interesting because it might affect transcription. Further genetic studies of Med15 are warranted.
HIC2 (hypermethylated in cancer, 2 [MIM 607712]) is particularly important to discuss with respect to distal 22q11.2 deletions. HIC2 maps to a 15 kb region of unique sequences that disrupts an internal part of LCR22D, near its 3′ end. The exact function of HIC2 is not known, but it is related to HIC1, which encodes a tumor suppressor. A recent study of Hic2 in mouse models demonstrated that it is sensitive to gene dosage for heart development.47 Although Hic2−/− embryos die by E10.5, some Hic2+/− mice show perinatal lethality.47 Of interest, a third of Hic2+/− embryos had a VSD with an overriding aorta.47 It is not known definitively whether HIC2 is hemizygously deleted in individuals that have chromosome endpoints within LCR22D (LCR22A–D, B–C, or C–D). Definitive proof would need to come from direct quantitative sequence analyses. Nonetheless, HIC2 is hemizygously deleted in individuals with deletions that include a deletion of LCR22D.47 Thus, haploinsufficiency of HIC2 might be an important contributor to that of individuals with distal chromosome 22q11.2 rearrangements.
Conclusions
In conclusion, our data strongly suggest that haploinsufficiency of CRKL can contribute to the etiology of CHD in individuals with nested distal deletions. The allelic series of altered Crkl expression in mice confirms the possibility that CRKL is dosage sensitive in humans. This work also suggests that CRKL might serve as a genetic modifier of cardiac anomalies in individuals with the typical 3 Mb, LCR22A–D deletion. Finally, haploinsufficiency of both CRKL and MAPK1 might result in a higher frequency of CHD. This would suggest that the CRKL-MAPK pathway might be important in human cardiac development and disease.
Acknowledgments
We would like to thank the families and individuals who kindly provided biological samples for genetic research as well as the information about their cardiac diagnosis. We also want to thank the Histopathology and Genomics Core Facilities at Einstein. This work was supported by NIH P01HD070454 (B.E.M., D.M.M.-M., B.S.E., E.G.) and an AHA postdoctoral grant 12POST9100003 (S.E.R.).
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Albert Einstein College of Medicine: HistoPathology, http://www.einstein.yu.edu/histopathology/page.aspx
Gene Expression Analysis Using TaqMan® Assays, http://www.lifetechnologies.com/us/en/home/life-science/pcr/real-time-pcr/real-time-pcr-assays/taqman-gene-expression.html
OMIM, http://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
References
- 1.van der Linde D., Konings E.E., Slager M.A., Witsenburg M., Helbing W.A., Takkenberg J.J., Roos-Hesselink J.W. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J. Am. Coll. Cardiol. 2011;58:2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Kodo K., Yamagishi H. A decade of advances in the molecular embryology and genetics underlying congenital heart defects. Circ. J. 2011;75:2296–2304. doi: 10.1253/circj.cj-11-0636. [DOI] [PubMed] [Google Scholar]
- 3.Burn J., Goodship J. Developmental genetics of the heart. Curr. Opin. Genet. Dev. 1996;6:322–325. doi: 10.1016/s0959-437x(96)80009-8. [DOI] [PubMed] [Google Scholar]
- 4.Edelmann L., Pandita R.K., Spiteri E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum. Mol. Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- 5.Shaikh T.H., Kurahashi H., Saitta S.C., O’Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Emanuel B.S. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 6.Rauch A., Zink S., Zweier C., Thiel C.T., Koch A., Rauch R., Lascorz J., Hüffmeier U., Weyand M., Singer H., Hofbeck M. Systematic assessment of atypical deletions reveals genotype-phenotype correlation in 22q11.2. J. Med. Genet. 2005;42:871–876. doi: 10.1136/jmg.2004.030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Miñaur S., Fantes J., Murray R.S., Porteous M.E., Strain L., Burns J.E., Stephen J., Warner J.P. A novel atypical 22q11.2 distal deletion in father and son. J. Med. Genet. 2002;39:E62. doi: 10.1136/jmg.39.10.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindsay E.A., Goldberg R., Jurecic V., Morrow B., Carlson C., Kucherlapati R.S., Shprintzen R.J., Baldini A. Velo-cardio-facial syndrome: frequency and extent of 22q11 deletions. Am. J. Med. Genet. 1995;57:514–522. doi: 10.1002/ajmg.1320570339. [DOI] [PubMed] [Google Scholar]
- 9.Carlson C., Sirotkin H., Pandita R., Goldberg R., McKie J., Wadey R., Patanjali S.R., Weissman S.M., Anyane-Yeboa K., Warburton D. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am. J. Hum. Genet. 1997;61:620–629. doi: 10.1086/515508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merscher S., Funke B., Epstein J.A., Heyer J., Puech A., Lu M.M., Xavier R.J., Demay M.B., Russell R.G., Factor S. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 11.Jerome L.A., Papaioannou V.E. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 12.Yagi H., Furutani Y., Hamada H., Sasaki T., Asakawa S., Minoshima S., Ichida F., Joo K., Kimura M., Imamura S. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 13.Zweier C., Sticht H., Aydin-Yaylagül I., Campbell C.E., Rauch A. Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions. Am. J. Hum. Genet. 2007;80:510–517. doi: 10.1086/511993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guris D.L., Fantes J., Tara D., Druker B.J., Imamoto A. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat. Genet. 2001;27:293–298. doi: 10.1038/85855. [DOI] [PubMed] [Google Scholar]
- 15.Verhagen J.M., Diderich K.E., Oudesluijs G., Mancini G.M., Eggink A.J., Verkleij-Hagoort A.C., Groenenberg I.A., Willems P.J., du Plessis F.A., de Man S.A. Phenotypic variability of atypical 22q11.2 deletions not including TBX1. Am. J. Med. Genet. A. 2012;158A:2412–2420. doi: 10.1002/ajmg.a.35517. [DOI] [PubMed] [Google Scholar]
- 16.Kurahashi H., Nakayama T., Osugi Y., Tsuda E., Masuno M., Imaizumi K., Kamiya T., Sano T., Okada S., Nishisho I. Deletion mapping of 22q11 in CATCH22 syndrome: identification of a second critical region. Am. J. Hum. Genet. 1996;58:1377–1381. [PMC free article] [PubMed] [Google Scholar]
- 17.Guris D.L., Duester G., Papaioannou V.E., Imamoto A. Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Dev. Cell. 2006;10:81–92. doi: 10.1016/j.devcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Moon A.M., Guris D.L., Seo J.H., Li L., Hammond J., Talbot A., Imamoto A. Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev. Cell. 2006;10:71–80. doi: 10.1016/j.devcel.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ten Hoeve J., Morris C., Heisterkamp N., Groffen J. Isolation and chromosomal localization of CRKL, a human crk-like gene. Oncogene. 1993;8:2469–2474. [PubMed] [Google Scholar]
- 20.Matsuda M., Tanaka S., Nagata S., Kojima A., Kurata T., Shibuya M. Two species of human CRK cDNA encode proteins with distinct biological activities. Mol. Cell. Biol. 1992;12:3482–3489. doi: 10.1128/mcb.12.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prosser S., Sorokina E., Pratt P., Sorokin A. CrkIII: a novel and biologically distinct member of the Crk family of adaptor proteins. Oncogene. 2003;22:4799–4806. doi: 10.1038/sj.onc.1206714. [DOI] [PubMed] [Google Scholar]
- 22.Birge R.B., Kalodimos C., Inagaki F., Tanaka S. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun. Signal. 2009;7:13. doi: 10.1186/1478-811X-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuda M., Tanaka S. Roles for crk in cancer metastasis and invasion. Genes Cancer. 2012;3:334–340. doi: 10.1177/1947601912458687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fathers K.E., Bell E.S., Rajadurai C.V., Cory S., Zhao H., Mourskaia A., Zuo D., Madore J., Monast A., Mes-Masson A.M. Crk adaptor proteins act as key signaling integrators for breast tumorigenesis. Breast Cancer Res. 2012;14:R74. doi: 10.1186/bcr3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park T.J., Boyd K., Curran T. Cardiovascular and craniofacial defects in Crk-null mice. Mol. Cell. Biol. 2006;26:6272–6282. doi: 10.1128/MCB.00472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rump P., de Leeuw N., van Essen A.J., Verschuuren-Bemelmans C.C., Veenstra-Knol H.E., Swinkels M.E., Oostdijk W., Ruivenkamp C., Reardon W., de Munnik S. Central 22q11.2 deletions. Am. J. Med. Genet. A. 2014;164A:2707–2723. doi: 10.1002/ajmg.a.36711. [DOI] [PubMed] [Google Scholar]
- 27.Xu X., Li C., Garrett-Beal L., Larson D., Wynshaw-Boris A., Deng C.X. Direct removal in the mouse of a floxed neo gene from a three-loxP conditional knockout allele by two novel approaches. Genesis. 2001;30:1–6. doi: 10.1002/gene.1025. [DOI] [PubMed] [Google Scholar]
- 28.Aihara K., Azuma H., Akaike M., Sata M., Matsumoto T. Heparin cofactor II as a novel vascular protective factor against atherosclerosis. J. Atheroscler. Thromb. 2009;16:523–531. doi: 10.5551/jat.1552. [DOI] [PubMed] [Google Scholar]
- 29.Kumar A., Bhandari A., Sarde S.J., Goswami C. Genetic variants and evolutionary analyses of heparin cofactor II. Immunobiology. 2014;219:713–728. doi: 10.1016/j.imbio.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Yu Q., Zhao Z., Sun J., Guo W., Fu J., Burnstock G., He C., Xiang Z. Expression of P2X6 receptors in the enteric nervous system of the rat gastrointestinal tract. Histochem. Cell Biol. 2010;133:177–188. doi: 10.1007/s00418-009-0659-0. [DOI] [PubMed] [Google Scholar]
- 31.Hausmann R., Bodnar M., Woltersdorf R., Wang H., Fuchs M., Messemer N., Qin Y., Günther J., Riedel T., Grohmann M. ATP binding site mutagenesis reveals different subunit stoichiometry of functional P2X2/3 and P2X2/6 receptors. J. Biol. Chem. 2012;287:13930–13943. doi: 10.1074/jbc.M112.345207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolf S., Janzen A., Vékony N., Martiné U., Strand D., Closs E.I. Expression of solute carrier 7A4 (SLC7A4) in the plasma membrane is not sufficient to mediate amino acid transport activity. Biochem. J. 2002;364:767–775. doi: 10.1042/BJ20020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaillancourt F.H., Brault M., Pilote L., Uyttersprot N., Gaillard E.T., Stoltz J.H., Knight B.L., Pantages L., McFarland M., Breitfelder S. Evaluation of phosphatidylinositol-4-kinase IIIα as a hepatitis C virus drug target. J. Virol. 2012;86:11595–11607. doi: 10.1128/JVI.01320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bojjireddy N., Botyanszki J., Hammond G., Creech D., Peterson R., Kemp D.C., Snead M., Brown R., Morrison A., Wilson S. Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J. Biol. Chem. 2014;289:6120–6132. doi: 10.1074/jbc.M113.531426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W., Sun T., Cao J., Liu F., Tian Y., Zhu W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp. Cell Res. 2012;318:944–954. doi: 10.1016/j.yexcr.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Nacak T.G., Leptien K., Fellner D., Augustin H.G., Kroll J. The BTB-kelch protein LZTR-1 is a novel Golgi protein that is degraded upon induction of apoptosis. J. Biol. Chem. 2006;281:5065–5071. doi: 10.1074/jbc.M509073200. [DOI] [PubMed] [Google Scholar]
- 37.Macfarlan T., Kutney S., Altman B., Montross R., Yu J., Chakravarti D. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. J. Biol. Chem. 2005;280:7346–7358. doi: 10.1074/jbc.M411675200. [DOI] [PubMed] [Google Scholar]
- 38.Macfarlan T., Parker J.B., Nagata K., Chakravarti D. Thanatos-associated protein 7 associates with template activating factor-Ibeta and inhibits histone acetylation to repress transcription. Mol. Endocrinol. 2006;20:335–347. doi: 10.1210/me.2005-0248. [DOI] [PubMed] [Google Scholar]
- 39.Côté F., Boisvert F.M., Grondin B., Bazinet M., Goodyer C.G., Bazett-Jones D.P., Aubry M. Alternative promoter usage and splicing of ZNF74 multifinger gene produce protein isoforms with a different repressor activity and nuclear partitioning. DNA Cell Biol. 2001;20:159–173. doi: 10.1089/104454901300069004. [DOI] [PubMed] [Google Scholar]
- 40.Gray P.A., Fu H., Luo P., Zhao Q., Yu J., Ferrari A., Tenzen T., Yuk D.I., Tsung E.F., Cai Z. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 41.Xie Q., Lin T., Zhang Y., Zheng J., Bonanno J.A. Molecular cloning and characterization of a human AIF-like gene with ability to induce apoptosis. J. Biol. Chem. 2005;280:19673–19681. doi: 10.1074/jbc.M409517200. [DOI] [PubMed] [Google Scholar]
- 42.Grondin B., Côté F., Bazinet M., Vincent M., Aubry M. Direct interaction of the KRAB/Cys2-His2 zinc finger protein ZNF74 with a hyperphosphorylated form of the RNA polymerase II largest subunit. J. Biol. Chem. 1997;272:27877–27885. doi: 10.1074/jbc.272.44.27877. [DOI] [PubMed] [Google Scholar]
- 43.Hwang J., Kalinin A., Hwang M., Anderson D.E., Kim M.J., Stojadinovic O., Tomic-Canic M., Lee S.H., Morasso M.I. Role of Scarf and its binding target proteins in epidermal calcium homeostasis. J. Biol. Chem. 2007;282:18645–18653. doi: 10.1074/jbc.M702035200. [DOI] [PubMed] [Google Scholar]
- 44.Anastasio N., Ben-Omran T., Teebi A., Ha K.C., Lalonde E., Ali R., Almureikhi M., Der Kaloustian V.M., Liu J., Rosenblatt D.S. Mutations in SCARF2 are responsible for Van Den Ende-Gupta syndrome. Am. J. Hum. Genet. 2010;87:553–559. doi: 10.1016/j.ajhg.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maerki S., Olma M.H., Staubli T., Steigemann P., Gerlich D.W., Quadroni M., Sumara I., Peter M. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J. Cell Biol. 2009;187:791–800. doi: 10.1083/jcb.200906117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beck J., Maerki S., Posch M., Metzger T., Persaud A., Scheel H., Hofmann K., Rotin D., Pedrioli P., Swedlow J.R. Ubiquitylation-dependent localization of PLK1 in mitosis. Nat. Cell Biol. 2013;15:430–439. doi: 10.1038/ncb2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dykes I.M., van Bueren K.L., Ashmore R.J., Floss T., Wurst W., Szumska D., Bhattacharya S., Scambler P.J. HIC2 is a novel dosage-dependent regulator of cardiac development located within the distal 22q11 deletion syndrome region. Circ. Res. 2014;115:23–31. doi: 10.1161/CIRCRESAHA.115.303300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


