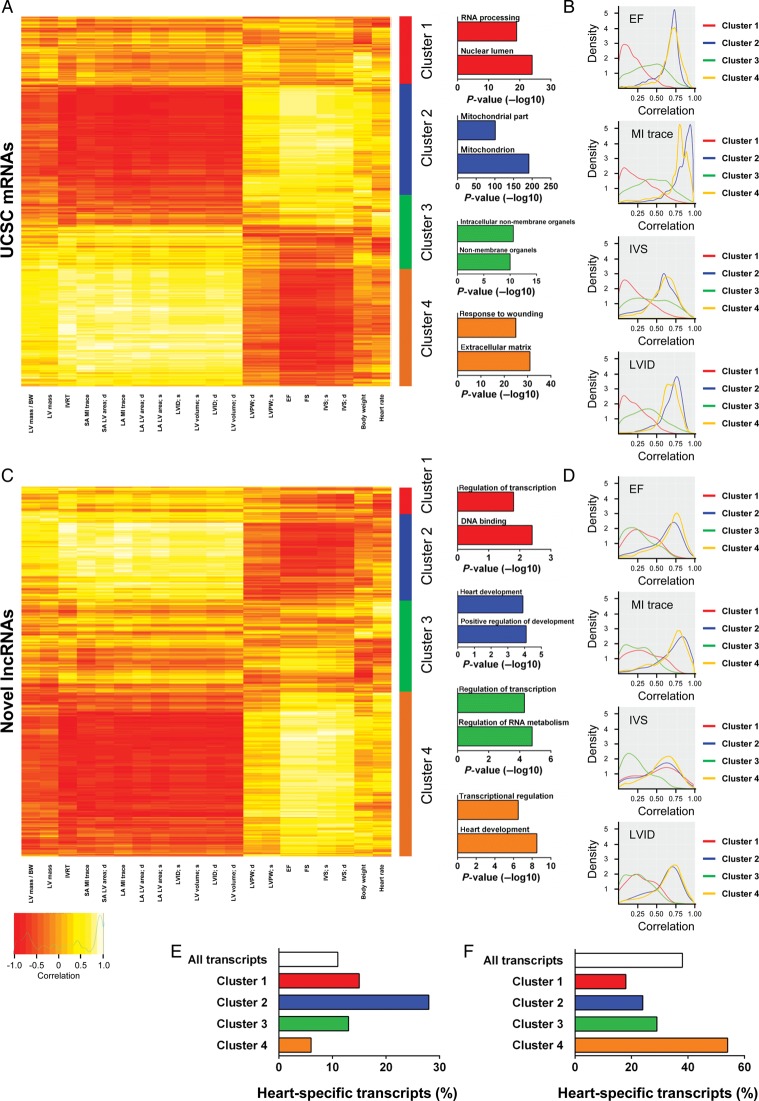
Figure 3.
Physiological clustering of the cardiac transcriptome. The cardiac transcriptome is highly correlated with cardiac physiological traits. (A) Clustering of UCSC mRNAs based on correlation of expression with echocardiography-derived physiological traits after myocardial infarction (Sham-operated and myocardial infarction samples were used for correlations). Clusters are illustrated right of the heatmap with top-enriched gene ontology terms for coding genes in each cluster displayed. (B) Kernel density plots of correlation of mRNA expression with selected physiological traits (ejection fraction, myocardial infarction trace, interventricular septal, and left ventricular internal diameter) within each cluster. (C) Clustering based on correlation of novel long non-coding RNA expression with echocardiography-derived physiological traits. Clusters are illustrated right of the heatmap with top-enriched terms for coding genes closest to novel long non-coding RNA. (D) Kernel density plots of correlation of long non-coding RNA expression with selected physiological traits (ejection fraction, myocardial infarction trace, interventricular septal, and left ventricular internal diameter) within each cluster. Heart specificity of UCSC mRNAs (E) and novel long non-coding RNAs (F) in clusters one to four.