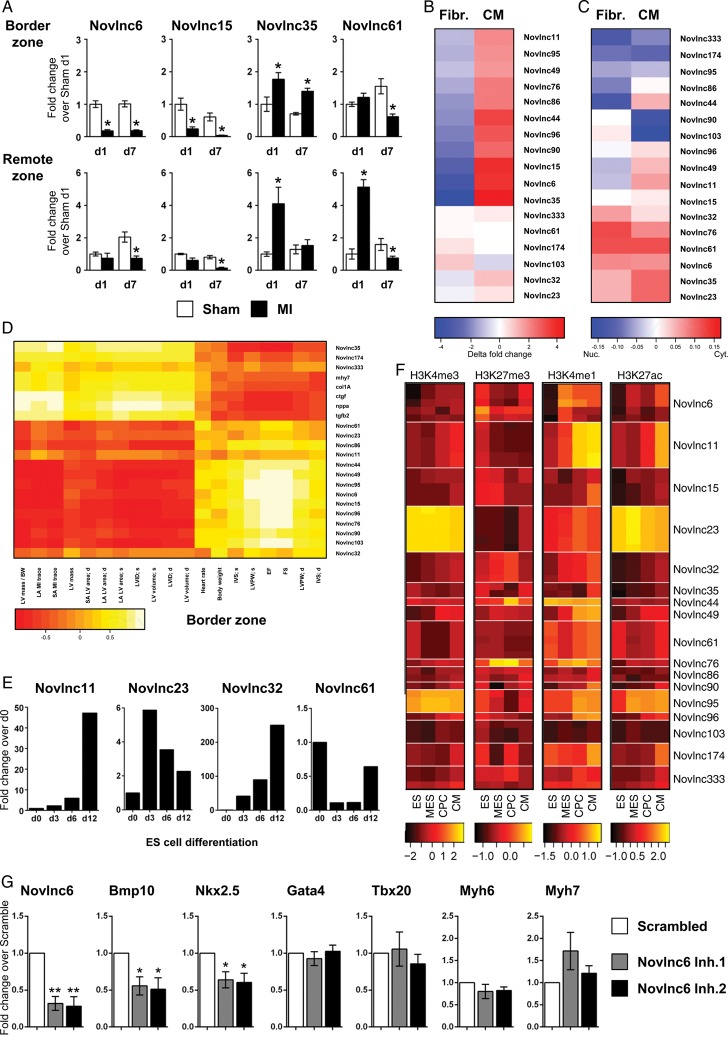
Figure 6.
Selection and validation of novel long non-coding RNAs. High priority candidates were selected based on their association with unique and specific characteristics. See Supplementary material online, Table S5 (A) Quantitative RT–PCR analysis of selected novel long non-coding RNAs in Sham-operated (white bar) and infarcted heart (black bar), in the border and remote zones at 1 and 7 days post-infarction. (B) Heatmap representation of qRT–PCR analysis of long non-coding RNAs in cardiomyocytes and fibroblasts isolated from neonatal mouse hearts. (C) Heatmap representation of qRT–PCR analysis of long non-coding RNAs in the cytoplasmic or nuclear fractions of cardiomyocytes and fibroblasts from neonatal mouse hearts. Blue indicates nuclear enrichment and red cytoplasmic enrichment. (D) Heatmap representation of correlation between validated novel long non-coding RNA expression in the border zone and echocardiography-derived physiological traits after infarction. (E) Expression of selected novel long non-coding RNA in embryonic stem, mesodermal precursors, cardiac precursor cell and cardiomyocyte as measured by qRT–PCR. (F) Heatmaps of ChIP enrichment values of different chromatin modification at TSS region of validated novel long non-coding RNA in embryonic stem, mesodermal precursors, cardiac precursor cell, and cardiomyocyte. Colour scale is based on log10 of the ChIP signal. (G) Mouse neonatal cardiomyocytes were transfected with GapmeRs targeting a novlnc6 or random scrambled sequence. Cells were harvested 48 h post-transfection and assayed for Novlnc6, Bmp10, Nkx2–5, GATA4, Tbx20, Myh6, and Myh7 expression by qRT–PCR. Bars represent means ± SEM (n = 6 independent experiments). **P < 0.001; *P < 0.05.