Figure 1.
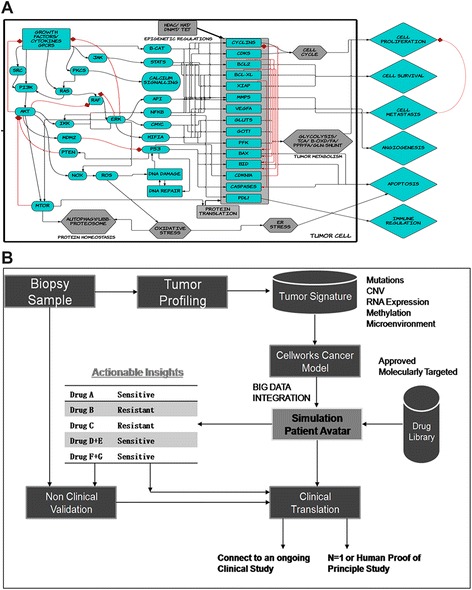
Predictive platform design and study workflow. (A) Schematic representing the crosstalk between various pathways in the plasma cell simulation platform. An oversimplified graphical representation of the signaling components, including key kinases, transcription factors and genes, that underlie various cancer phenotypes incorporated in the network is presented. The schematic also highlights cellular processes that are modeled, including epigenetic regulation, tumor metabolism, oxidative stress, protein homeostasis (proteasome and autophagy), cell cycle machinery and DNA repair pathways. The crosstalk between all these pathways represents our control non-triggered simulation platform that can be transitioned into the respective disease state by triggering the mutations and aberrations reported for a profile. (B) Description of process for designing personalized therapy: The process flow for the N = 1 personalized therapy design as described in the Methods section is presented. A bone marrow sample from the patient tumor is profiled, and the tumor signature consisting of CNV, mutations and other relevant data is input into the simulation model to create the simulation patient avatar. A drug library is tested on this patient avatar to identify drugs and therapies predicted to be sensitive or resistant in the particular profile. This information can be validated ex vivo via non-clinical validation or clinically translated by either connecting to a specific ongoing clinical study or the personalized N = 1 option for that particular patient.
