Figure 2.
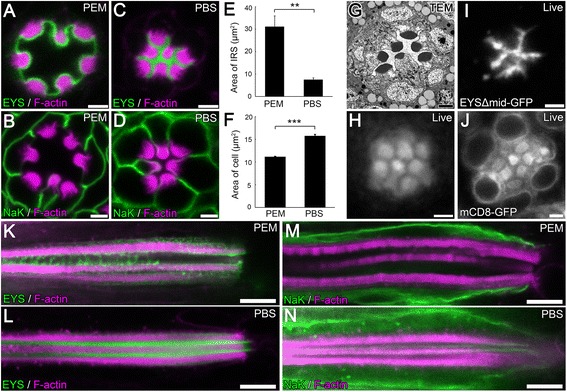
A severe distortion in lumen and cell morphology results when tissue is fixed in PEM (PIPES, EGTA, and MgSO 4 ) buffer. (A–D, K–N) Immunofluorescence micrographs of 96 hours (h) after puparium formation (APF) w 1118 (wild type) Drosophila ommatidium in a cross optical section (A–D) or in a vertical optical section (K–N) showing the positioning of the rhabdomeres and the size of the inter-rhabdomeral space (IRS). Tissues were fixed in PEM buffer (A,B,K,M) or in PBS buffer (C,D,L,N). The rhabdomeres, F-actin, are labeled with Phalloidin (magenta), and EYS (A,C,K,L) or Na+ K+ ATPase (NaK) which labels the basolateral membranes (B,D,M,N) are shown in green. (E–F) Quantitative analysis of the area of the IRS (E) or the average area of R1–R7 photoreceptor cells (F) in PEM or PBS buffered conditions as seen in (A–D). Values represent mean ± SEM. **P < 0.01, and ***P < 0.001. n = 3 retinas and in each retina, three ommatidia were quantified. (G) Transmission electron microscopy image of a 96 h APF w 1118 ommatidium showing the positioning of the rhabdomeres and the size of the IRS. (H) Bright field microscopy image of the ommatidium of a living 96 h APF w + wild type pupa, showing the positioning of the rhabdomeres. The rhabdomeres have higher reflection ability thus are brighter in the image, while the IRS and the cell bodies are darker. (I–J) Live, non-fixed retina tissue of 96 h APF pupae freshly dissected in Drosophila S2 cell medium. (I) EYSΔMid-GFP labels the IRS. (J) mCD8-GFP labels the plasma membrane of the photoreceptor cell, intracellular membrane structures, as well as microvillar membranes of the rhabdomeres. mCD8-GFP does not label structures in the nucleus. Scale bar, (A–D, G–J) 2 μm; (K–N) 5 μm.
