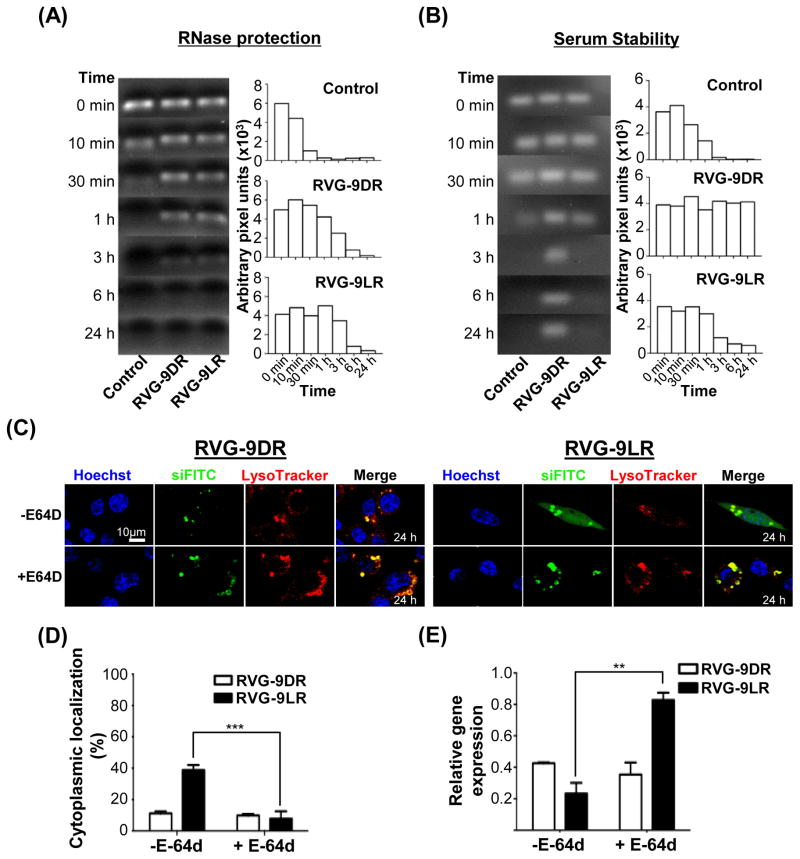
Figure 6. Endosomal proteolysis is required for siRNA release from late endosomes.
(A and B) Stability of RVG-9D/LR-siRNA complexes after exposure to RNase A (A) and 50% human AB serum (B) for the indicated periods of time. The bar graphs represent corresponding band intensities estimated as arbitrary pixel units using the Image J software. Control refers to siRNA treated identically in the absence of ligand-9R. (C and D) Fluorescence distribution in E-64d-treated Neuro2a cells 24h after exposure to RVG-9D/LR:siFITC. Nuclei were stained with Hoechst 33342 (blue) and endosomal vesicles with LysoTracker (red). Data in (D) were calculated using the Volocity software and a sample size of 50 cells. (E) Analysis of murine SOD-1 mRNA levels in Neuro2a cells 36h after exposure to RVG9D/LR-siSOD1 (100pmol siRNA, 10:1 peptide:siRNA). Error bars indicate SEM. **P<0.01, ***P<0.001.