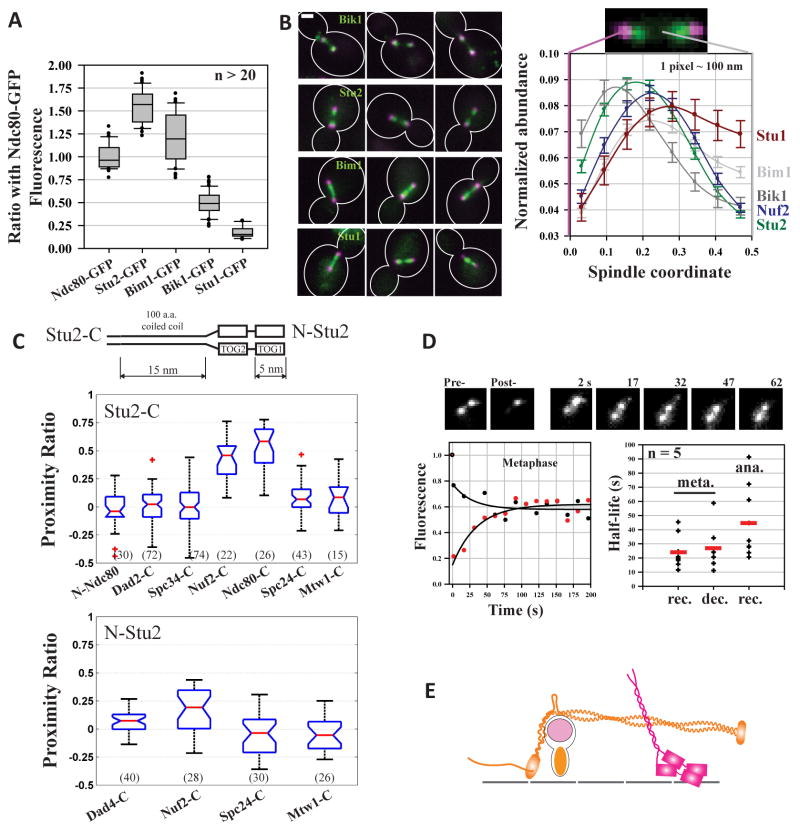
Figure 5. Abundance and spindle distribution of MAPs.
(A) Relative abundance of Stu2, Bik1, Bim1, and Stu1. In comparison with Stu2, other MAPs have a significantly lower abundance (p-value < 1e-9 from a two-sided Student’s t-test) and a significantly larger cell-to-cell variation in abundance (p-value < 0.02 from a two-sided F-test). (B) Representative micrographs of MAP distribution (spindle extremities marked by Spc97-mCherry, a spindle pole body protein; scale bar ~ 1 μm). Normalized distributions of Bik1, Bim1, Stu1, Stu2, and Nuf2 in metaphase arrested cells (mean +\− s.e.m.). (C) Top: schematic of the Stu2 dimer. Stu2 length was estimated by adding 5 nm length of its two TOG domains [42], and the estimated 15 nm contour length of its 100 amino acid long α-helical coiled coil domain (3.6 residues per turn and a pitch of 0.54 nm). FRET quantification for Stu2-C (upper graph) or N-Stu2 (lower graph) and kinetochore subunits. (D) Fluorescence Recovery after Photobleaching (FRAP) of Stu2-GFP. Red circles display the fluorescence recovery, and black circles display the concurrent fluorescence decay of the unbleached cluster in the same cell. The initial, steep decrease in the intensity of the unbleached cluster is due to inadvertent photobleaching. Black lines display single exponential fit of the data. The scatter plot displays the half-life for fluorescence recovery (rec.) and decay (dec.) in metaphase and only recovery in anaphase. Red line indicates the mean value. (E) Stu2 localization in the metaphase kinetochore.