Significance
We identified hypoxia-inducible domain family, member 1A (Higd1a) as a positive regulator of cytochrome c oxidase (CcO). CcO, the terminal component of the mitochondrial electron transfer system, reductively converts molecular oxygen to water coupled to pump protons across the inner mitochondrial membrane. Higd1a is transiently induced under hypoxic conditions and increases CcO activity by directly interacting with CcO in the vicinity of its active center. Induction of Higd1a leads to increased oxygen consumption and subsequent mitochondrial ATP synthesis, thereby improving cell viability under hypoxia.
Keywords: cytochrome c oxidase, oxidative phosphorylation, resonance Raman spectroscopy, ATP, oxygen
Abstract
Cytochrome c oxidase (CcO) is the only enzyme that uses oxygen to produce a proton gradient for ATP production during mitochondrial oxidative phosphorylation. Although CcO activity increases in response to hypoxia, the underlying regulatory mechanism remains elusive. By screening for hypoxia-inducible genes in cardiomyocytes, we identified hypoxia inducible domain family, member 1A (Higd1a) as a positive regulator of CcO. Recombinant Higd1a directly integrated into highly purified CcO and increased its activity. Resonance Raman analysis revealed that Higd1a caused structural changes around heme a, the active center that drives the proton pump. Using a mitochondria-targeted ATP biosensor, we showed that knockdown of endogenous Higd1a reduced oxygen consumption and subsequent mitochondrial ATP synthesis, leading to increased cell death in response to hypoxia; all of these phenotypes were rescued by exogenous Higd1a. These results suggest that Higd1a is a previously unidentified regulatory component of CcO, and represents a therapeutic target for diseases associated with reduced CcO activity.
Cytochrome c oxidase (CcO) (ferrocytochrome c: oxygen oxidoreductase, EC 1. 9. 3. 1) is the terminal component of the mitochondrial electron transfer system. CcO couples the oxygen-reducing reaction with the process of proton pumping. Aerobic organisms use this reaction to form a proton gradient across the mitochondrial inner membrane, which is ultimately used by the FoF1-ATP synthase to produce ATP.
Mammalian CcO is composed of 13 different subunits (1) containing four redox-active metal centers, two copper sites, and two heme a groups. These active centers accept electrons from cytochrome c and sequentially donate them to dioxygen. Our group and others have extensively analyzed the link between the oxygen reduction process and proton pumping at the active centers using crystallography, resonance Raman spectroscopy, and Fourier transform infrared spectroscopy (2–4). The metal ions in the copper sites and heme groups in the active centers are individually coordinated by the surrounding amino acids. We have shown that changes in the redox state cause 3D structural changes around the active centers, which in turn leads to alteration of the proton pump mediated by specific amino acid chains that coordinate each metal group (5). Thus, binding of an allosteric regulator close to the active centers might change the efficiency of both electron transfer to oxygen and proton pumping.
Several proteins involved in oxygen supply or metabolism are transcriptionally regulated by intracellular oxygen concentration: vascular endothelial growth factor (VEGF) (6), erythropoietin (EPO) (7), and G0/G1 switch gene 2 (G0s2) for FoF1-ATP synthase, as we recently revealed (8). Because CcO is the only enzyme in the body that can use oxygen for energy transduction, it has been suggested that the regulatory mechanism of CcO is dependent on oxygen concentration (9–12); however, this has yet to be demonstrated. In this study, we aimed to identify a regulator of CcO driven by low oxygen concentration.
In this study, by screening for hypoxia-inducible genes, we discovered that hypoxia inducible domain family, member 1A (Higd1a) is a positive regulator of CcO. Furthermore, using our recently established ATP-sensitive fluorescence resonance energy transfer (FRET) probe, we demonstrated that Higd1a increased mitochondrial ATP production. We also showed that Higd1a directly bound CcO and changed the structure of its active center.
Results
Higd1a Expression Is Induced Early in the Response to Hypoxia.
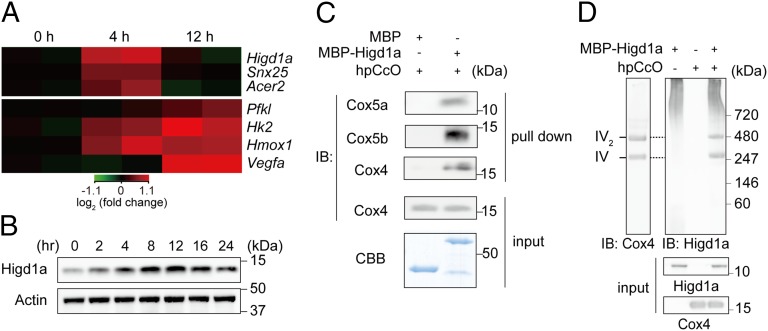
During the first few hours of hypoxia, CcO and oxidative phosphorylation (OXPHOS) activity is activated, presumably to fully use any remaining oxygen (12). At later time points, metabolism shifts toward glycolysis. Therefore, we hypothesized that a positive regulator of CcO must be up-regulated during an early stage of hypoxia, but down-regulated when glycolysis-related genes become elevated. To identify early hypoxia responsive genes that might regulate CcO activity, we analyzed gene-expression profiles of neonatal rat cardiomyocytes, one of the most mitochondria-rich cell types, exposed to hypoxic conditions (1% oxygen for 0, 4, or 12 h). Focusing on the genes whose expression was induced more than two-fold at 4 h relative to the prestimulation stage, but then decreased by 12 h, we identified three genes (Fig. 1A and Fig. S1 A and B). Next, we prioritized genes that were (i) well conserved among eukaryotes and (ii) listed in MitoCarta (13); only one gene, Higd1a, satisfied both criteria. To analyze the endogenous expression levels of Higd1a in rat cardiomyocytes, we raised a specific antibody against Higd1a and confirmed its specificity (Fig. S2 A and B). In cardiomyocytes exposed to hypoxia, Higd1a protein levels increased gradually from 0 to 12 h and then decreased by 24 h (Fig. 1B). Immunofluorescence revealed that both endogenous and exogenous Higd1a localized in the mitochondria (Fig. S2C).
Fig. 1.
Hypoxia-inducible Higd1a directly binds to highly purified cytochrome c oxidase (hpCcO). (A) Heat map of three genes (Upper) identified as relatively rapid and transiently induced in response to hypoxia in rat neonatal cardiomyocytes, compared with genes known to be hypoxia inducible (Pfkl, Hk2, Hmox1, and Vegfa) (Lower). (B) Expression of the Higd1a protein was elevated in response to hypoxia. (C) In vitro pull-down assay with amylose resin revealed direct binding between MBP-Higd1a and the hpCcO from bovine heart. Loading controls for the hpCcO and MBP-fusion proteins are shown in immunoblots for anti-CcO subunits and CBB staining, respectively. (D) MBP-Higd1a directly integrates into hpCcO. Mixed MBP-fusion proteins and hpCcO containing 0.2% n-decyl-β-d-maltoside (DM) were resolved by blue native PAGE (BN-PAGE), followed by immunoblotting with anti-Cox4 to detect CcO and anti-Higd1a to detect Higd1a.
Higd1a Directly Integrates into the CcO Macromolecular Complex.
Because Rcf1a, the yeast homolog of Higd1a, associates with CcO (9–11), we first tested whether mammalian Higd1a binds to CcO in vivo. Indeed, endogenous binding between Higd1a and CcO in rat cardiomyocytes was confirmed by immunocapture with an anti-Higd1a antibody (Fig. S3A) and verified by reciprocal coimmunoprecipitation with an anti-Cox4 antibody (Fig. S3B). This in vivo interaction was further validated by blue native PAGE (BN-PAGE) of mitochondrial fractions from rat cardiomyocytes (Fig. S3C).
Because preparation of the CcO macromolecular complex, which consists of 13 subunits, is technically demanding, it has remained unclear whether Rcf1a/Higd1a binding to CcO is direct. To address this issue, we performed an in vitro pull-down assay using highly purified bovine CcO (hpCcO), which we prepared by dissolving microcrystals used for X-ray structural analysis (14). Notably, recombinant maltose binding protein-fused bovine Higd1a (MBP-Higd1a) (Fig. S4) directly associated with hpCcO (Fig. 1C). Furthermore, to assess macromolecular complex formation, we performed BN-PAGE followed by immunoblotting with an antibody against Higd1a, demonstrating that recombinant Higd1a indeed integrated into hpCcO (Fig. 1D). With these results, we conclude that Higd1a directly associates and integrates into the CcO macromolecular complex.
Higd1a Causes Structural Changes in CcO and Influences the Active Center of Heme a.
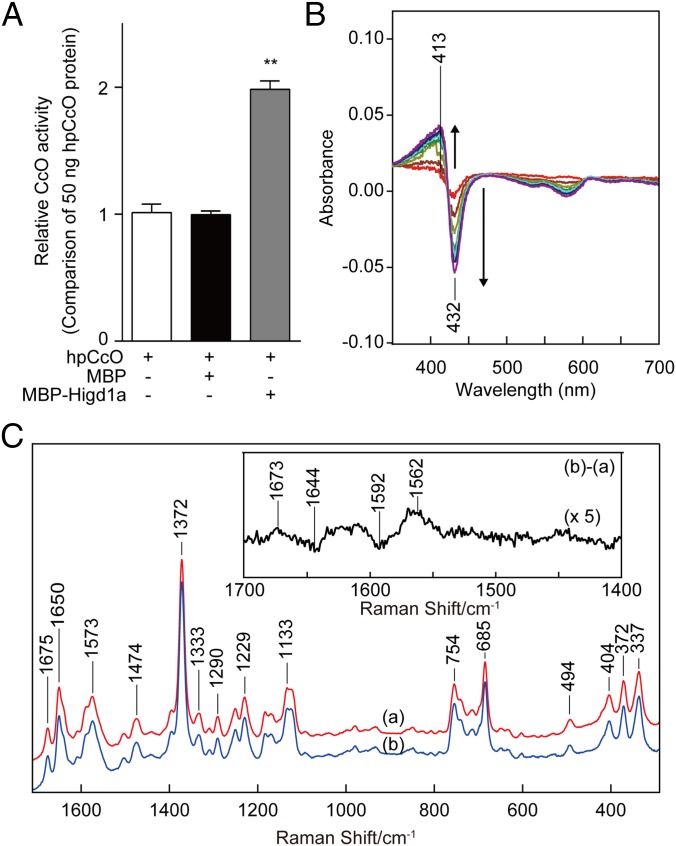
To explore the relevance of the interaction between Higd1a and CcO, we investigated whether recombinant Higd1a affects hpCcO enzymatic activity. Strikingly, direct addition of MBP-Higd1a to hpCcO significantly increased CcO activity to twice that of hpCcO alone or hpCcO mixed with MBP (Fig. 2A). This significant increase in hpCcO activity stimulated by Higd1a led us to speculate that Higd1a causes a structural change at the active centers of CcO.
Fig. 2.
Higd1a regulates CcO activity through the structural change of the active center in CcO. (A) CcO activity of hpCcO and hpCcO with either recombinant MBP or recombinant MBP-Higd1a. MBP-Higd1a causes an increase in CcO activity by almost twofold. Data represent the means ± SEM of five individual experiments. **P < 0.01, compared with MBP. (B) The difference in absorption spectra between MBP-Higd1a and oxidized hpCcO. MBP-Higd1a caused spectral changes at 413 and 432 nm. Intensity changes of oxidized hpCcO spectra are plotted at 1 min (red), 5 min (brown), 10 min (dark yellow), 15 min (green), 20 min (light blue), 25 min (blue), and 30 min (purple) after adding MPB-Higd1a. (C) Resonance Raman spectra of oxidized hpCcO at 0–5 min [spectrum (a)] and oxidized hpCcO mixed with MBP-Higd1a at 0–5 min [spectrum (b)]. The Inset shows the difference of the spectra [(b) − (a)].
Therefore, we next investigated whether Higd1a changes the intensity of the visible part of the absorption spectrum of oxidized CcO. MBP alone, used as a negative control, did not cause a significant change in the absorption spectra (Fig. S5). By contrast, MBP-Higd1a caused significant spectral changes at 413 nm and 432 nm (Fig. 2B), wavelengths that reflect conformational changes around the hemes in oxidized CcO (15).
To obtain further structural insights, we performed resonance Raman spectroscopy, a powerful and sensitive method for detecting kinetic structural changes that cannot be assessed by X-ray crystal structural analysis. Fig. 2C depicts the resonance Raman spectra of CcO with and without MBP-Higd1a, focusing on the heme structure by using 413 nm excitation. The resonance Raman band at 1,372 cm−1 in (a: hpCcO) and (b: hpCcO + Higd1a) is assignable to the ν4 mode of heme and is indicative of ferric heme. After the addition of recombinant Higd1a, the resonance Raman spectra demonstrated two sets of different peaks (or band shifts) at 1,562/1,592 cm−1 (the ν2 mode; a marker for the spin state of heme) (16) and 1,673/1,644 cm−1 (the νCH = O mode of the formyl group of heme a) (17). Importantly, the frequency shift of the band at 1,592 cm−1 to 1,562 cm−1 is attributable to partial conversion of heme from a low-spin to a high-spin state. In oxidized CcO, only heme a includes low-spin iron; therefore, heme a, but not heme a3, is responsible for the band shift (16). These data suggest that the binding of Hig1a to CcO caused structural changes at heme a, the active center of CcO.
Higd1a Positively Regulates CcO Activity and Subsequent Mitochondrial OXPHOS.
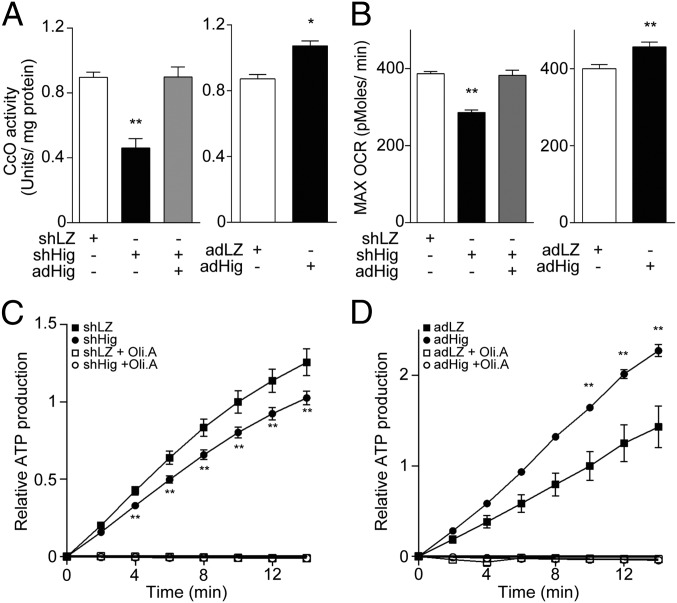
Next, we investigated whether Higd1a truly regulates CcO activity in vivo. To this end, we assessed biochemical CcO activity in rat cardiomyocytes with modified expression of Higd1a. Notably, we observed a significant decrease in CcO activity in Higd1a knockdown cells. This effect was rescued by overexpression of Higd1a, eliminating the possibility of off-target effects in the RNAi experiment (Fig. 3A, Left). Moreover, overexpression of Higd1a alone increased the basal CcO activity (Fig. 3A, Right). These data suggest that Higd1a is an endogenous and positive regulator of CcO.
Fig. 3.
Higd1a positively modulates mitochondrial respiration by altering CcO activity. (A, Left) Mitochondrial fraction from rat cardiomyocytes expressing shLacZ (shLZ), shHigd1a (shHig), or both shHig and adHigd1a (adHig) were subjected to the CcO activity assay. (Right) CcO activity was measured in cardiomyocytes treated with either adLacZ (adLZ) or adHig. Data represent the means of four individual experiments. (B, Left) The maximum oxygen consumption rate (max OCR) in rat cardiomyocytes transfected with the indicated adenovirus was measured after treatment with oligomycin A and fluorocarbonyl cyanide phenylhydrazone (FCCP). Knockdown of Higd1a resulted in a significant decrease in max OCR, which was rescued by exogenously expressed Higd1a. (Right) Overexpression of Higd1a significantly increased max OCR compared with the cells with adLZ (n = 20 for each group). (C) The relative ATP production rate of cardiomyocytes treated with shLZ or shHig was measured by the MASC assay (n = 6). A numerical value of ATP production at 10 min in shLZ groups is regarded as 1.0. (D) The relative ATP production rate of cardiomyocytes treated with adLZ or adHig was measured by MASC assay (n = 5). A numerical value of ATP production at 10 min in adLZ groups is regarded as 1.0. Data represent the means ± SEM; *P < 0.05, **P < 0.01.
To assess the effect of Higd1a on cellular respiration, we continuously measured the oxygen consumption rate (OCR) using a XF96 Extracellular Flux Analyzer (Seahorse Bioscience). Knockdown of Higd1a caused a significant decrease in both basal (Fig. S6A, Left) and maximum OCR, and these effects were rescued by exogenous expression of Higd1a (Fig. 3B, Left). Moreover, overexpression of Higd1a significantly increased both basal and maximum OCR (Fig. S6A, Right and Fig. 3B, Right).
Because the electron transport chain creates a proton gradient that drives FoF1-ATP synthase (complex V), ATP production is the overall outcome of mitochondrial OXPHOS. To determine whether modulation of CcO activity by Higd1a affects ATP production, we performed the mitochondrial activity of streptolysin O permeabilized cells (MASC) assay, a sensitive means of measuring the mitochondrial ATP production rate in semi-intact cells (18). Indeed, Higd1a knockdown caused a significant decrease in the ATP production rate relative to the control (Fig. 3C), whereas overexpression of Higd1a increased it (Fig. 3D). These results suggest that Higd1a modulates mitochondrial OXPHOS through CcO.
Higd1a Protects Cardiomyocytes Under Hypoxic Conditions by Increasing ATP Production.
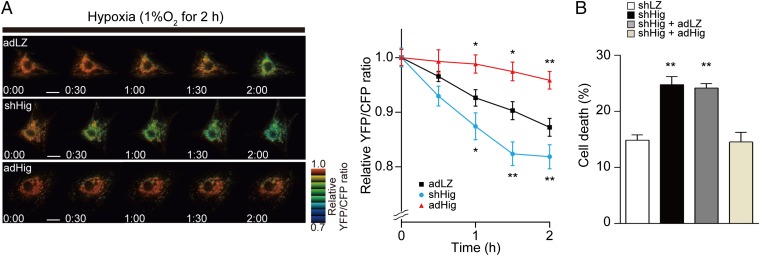
We reasoned that endogenous induction of Higd1a by hypoxia serves to maintain ATP production in mitochondria to the greatest extent possible when oxygen supply is limited. The intramitochondrial matrix ATP concentration ([ATP]mito) reflects mitochondrial ATP production far more sensitively than the cytosolic ATP concentration (8). Therefore, we next assessed the effect of Higd1a on ATP production in living cells using the FRET-based mitochondrial ATP biosensor Mit-ATeam (19). First, we examined the effect of KCN, an inhibitor of CcO. KCN significantly reduced the [ATP]mito (Fig. S7), suggesting that Mit-ATeam provides an effective means to monitor the functional consequences of changes in CcO activity. We then confirmed that hypoxia caused a gradual decline in [ATP]mito. Overexpression of Higd1a alleviated the decline in [ATP]mito during hypoxia, whereas knockdown of Higd1a accelerated the decrease in [ATP]mito relative to the control (Fig. 4A).
Fig. 4.
(A) Representative sequential YFP/CFP ratiometric images of Mit-ATeam fluorescence in cardiomyocytes expressing corresponding adenovirus during hypoxia (n = 12 for adLZ, n = 23 for shHig, n = 18 for adHig). All of the measurements were normalized to the ratio at time 0 and compared between adLZ and adHig or shHig. (Scale bar, 20 μm.) (B) Cell death of cardiomyocytes treated with shHig was significantly increased compared with the control, which was rescued by addition of adHig under hypoxic conditions for 24 h (n = 12 for each group). Data represent the means of three independent cultures, ± SEM; *P < 0.05, **P < 0.01, compared with control (adLZ or shLZ).
The yeast homolog Rcf1 plays a role in respiratory supercomplex stability, and the same is true for Higd2a, but not Higd1a (11). We investigated whether Higd1a affects respiratory supercomplex stability Higd1a-knockdown or -overexpressing cells. As shown in Fig. S8, there was no significant change in the abundance or composition of the respiratory supercomplex, suggesting that the effect of Higd1a described above is not a result of changes in supercomplex stability.
Finally, to test whether the effects of Higd1a on mitochondrial ATP synthesis affected overall cell viability, we analyzed the viability of cardiomyocytes subjected to hypoxia. Under hypoxic conditions, Higd1a-knockdown cells showed a significant increase in cell death, and this effect was rescued by exogenous expression of Higd1a (Fig. 4B and Fig. S9A). In addition, overexpression of Higd1a alone increased cellular tolerance to hypoxia (Fig. S9B). On the basis of these findings, we conclude that Higd1a positively regulates CcO activity and subsequently increases mitochondrial ATP production, thereby protecting cardiomyocytes against hypoxia.
Discussion
In this study, we demonstrated that recombinant Higd1a produced in Escherichia coli was incorporated into CcO complex purified from bovine heart. The data suggest that Higd1a directly bound to the already assembled CcO complex and increased its activity. Together with the fact that Higd1a expression was rapidly increased by hypoxia, this observation indicated that Higd1a is a positive regulator of CcO that preserves the proton-motive force under hypoxic cellular stress. Physiologically, Higd1a preserved ATP production in healthy cardiomyocytes under hypoxic conditions, which protected them from an energy crisis leading to cell death.
We demonstrated that Higd1a incorporated into the CcO complex and increased its activity. It remains unclear which part of CcO is essential for this change. Higd1a binding may affect the interaction of cytochrome c with CcO, modulate internal electron/proton transfer, or modify Kd/Km for O2 binding to CuB/heme a3. In fact, the resonance Raman spectroscopy experiment provided us with a clue to this question. First, we discovered that Higd1a markedly shifted the maximum Soret peak around 413 nm absorption, suggesting the occurrence of structural changes in heme that are usually observed during the reduction and oxidation process of CcO. This shift in absorbance prompted us to perform resonance Raman analysis at 413 nm excitation, a powerful tool for investigating the structure of heme and its vicinity. Higd1a induced a frequency shift of the band at 1,592 cm−1 to 1,562 cm−1 and 1,673/1,644 cm−1; the former frequency is attributed to partial conversion of heme from a low-spin to a high-spin state. In oxidized CcO, only heme a includes low-spin iron (16); therefore, heme a, but not heme a3, is responsible for this band shift.
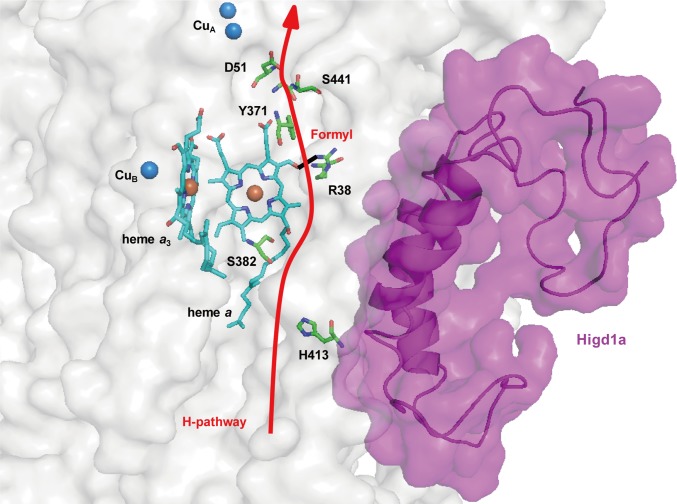
X-ray structural and mutational analyses for bovine heart CcO have demonstrated that protons are pumped through the hydrogen-bond network across the CcO molecule, designated the H pathway, located near heme a (20). The driving force for active proton transport is electrostatic repulsion between the proton in the hydrogen-bond network and the net positive charge of heme a. One of the critical sites for repulsion is the formyl group of heme a, which is hydrogen bonded to Arg38 of the CcO subunit I (21). In our study, resonance Raman spectroscopy revealed specific band shifts from 1,644 cm−1 to 1,673 cm−1, which can be attributed to the vibration of the formyl group of heme a. This observation suggests that Higd1a binding causes structural changes, particularly around heme a, weakening the hydrogen bond between the formyl group and Arg38 of the CcO subunit I, thereby leading to the acceleration of proton pumping efficiency (22). Thus, both band shifts suggest that structural change occurs in the vicinity of heme a rather than a3.
Following the resonance Raman analysis, we sought to determine the Higd1a-CcO binding site via simulation with the COOT software (23), using the previously reported structures of CcO (14) and Higd1a (24). From our structural analysis, CcO contains a cleft composed of relatively few protein subunits near the active centers (Fig. S10A). Notably, Higd1a was predicted to integrate into the cleft of CcO near heme a and Arg38 (Fig. S10), consistent with the results of the resonance Raman analysis. Thus, it is likely that Higd1a bound to the cleft of CcO, leading to swift structural change around heme a and Arg38 and accelerating the proton-pumping H pathway, thereby increasing CcO activity (Fig. 5). Furthermore, when we retrospectively reviewed the purification process of the CcO complex, comprising 13 subunits from the bovine heart, we found that Higd1a remained associated with CcO up to the final step, which required detergent exchange (14). This led us to speculate that Higd1a represents a 14th identified subunit of CcO that is endogenously induced by hypoxia and integrates into the open cleft of CcO to positively regulate its activity. Although the resonance Raman data and docking model simulation are consistent with the idea that Higd1a binding causes structural change around heme a, these data are limited because of their speculative nature. Therefore, to confirm these findings, we are currently trying to crystallize the CcO-Higd1a complex to reveal the conformational changes of CcO, particularly around the heme a site.
Fig. 5.
Higd1a acts on the H pathway. Model depicting our docking simulation (side view) and its relationship with the H pathway. The model shows the location of Higd1a (magenta) in the CcO complex (white) and its relationship to R38 of cytochrome c oxidase subunit I and the formyl group of heme a, a component of the H pathway (red arrow).
Higd1a was originally identified as a mitochondrial inner membrane protein whose expression is induced by hypoxia (25). Higd1a augments cell survival under hypoxic stress in pancreatic cells (26), and it exerts its protective effect by induction of mitochondrial fission (27). The precise relationship between these reports and our data is not clear. However, our results suggest that the elevation of CcO activity by Higd1a preserves the proton-motive force, which is prerequisite for mitochondria function, thereby leading to increased mitochondrial fission and/or the prevention of apoptosis.
The existence of a direct CcO allosteric activator suggests that there is a structural basis for the intrinsic activation in the CcO complex. To explore this idea further, a screen for small compounds that simply increase the activity of highly purified CcO in vitro has been initiated. Compounds that mimic the effect of Higd1a can preserve ATP production even under hypoxic condition, and hence are expected to exert cellular protective effects particularly when OXPHOS activity is reduced. Recent work showed that lowering the activity of OXPHOS causes the cellular senescence (28), diabetes mellitus (29), and neurodegenerative diseases (30). In addition, several currently intractable mitochondrial diseases are caused by mutations in mitochondrial genes or nuclear genes that lead to dysfunction in mitochondrial OXPHOS. Notably, decreased CcO activity is most frequently observed among patients with mitochondrial diseases (31). Therefore, small compounds that mimic the effect of Higd1a will have therapeutic potential for various acute and chronic diseases including ischemic, metabolic, and mitochondrial diseases.
Materials and Methods
Purification of Recombinant Higd1a Protein.
The full-length bovine Higd1a cDNA was purchased from GE Healthcare. Then the coding sequence of bovine Higd1a was cloned in-frame with an ATG start codon, in the pET21a expression vector (Novagen for overexpression in E. coli). A MBP was fused in-frame at the amino terminus for purification. The resulting plasmid was transformed into BL21-Star (DE3; Invitrogen), and the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside caused the expression of MBP-Higd1a protein. The cells were sonicated and solubilized by 1% n-decyl-β-d-maltoside (DM). The recombinant protein was purified with amylose resin (New England Biolabs), and eluted by 20 mM maltose (pH 6.8 or 8.0, 100 mM sodium phosphate buffer containing 0.2% DM). The eluted protein was concentrated and maltose removed using Amicon Ultra-0.5 10K (Millipore).
Resonance Raman Spectroscopy.
Absorption spectra of the samples were measured by a spectrophotometer (Hitachi, U3310) with the path length of 2 mm in 100 mM sodium phosphate buffer (pH 8.0) containing 0.2% DM. The reaction mixture was measured immediately and spectra were recorded every 5 min for 30 min. The protein concentration was 8 μM.
Raman scattering of the samples were measured in a cylindrical spinning cell with excitation at 413.1 nm with a Kr+ laser (Spectra Physics, model 2060), and the incident power was 500 μW. The detector was a liquid N2-cooled CCD detector (Roper Scientific, Spec-10: 400B/LN). Raman shifts were calibrated with indene as the frequency standard. Raman spectrum was divided by the “white light” spectrum that was determined by measuring the scattered radiation of an incandescent lamp by a white paper to compensate for the sensitivity difference of each CCD pixel and transmission curve of the notch filter to reject Rayleigh scattering. The accuracy of the peak position of well-defined Raman bands was ±1 cm−1. The protein concentration was 20 μM, and the reaction mixture was incubated for 30 min, before Raman measurements.
Measurement of CcO Activity.
CcO activity was measured spectrophotometrically (Shimazu, UV-2450) using a cytochrome c oxidase activity kit (Biochain). A total of 25 μg of mitochondrial pellets from cardiomyocytes was lysed with 1% n-dodecyl-β-d-maltoside (DDM), and subjected to measurement according to the manufacturer’s instructions (32). Concentrations of reduced/oxidized cytochrome c were determined using the extinction coefficient at 550 nm of 21.84 mM−1cm−1. For in vitro measurement, cytochrome c (Sigma) was reduced by ascorbic acid (Wako). Recombinant MBP-Higd1a (20 μM) and hpCcO (20 μM) were incubated at 25 °C for 30 min in the presence of 0.2% DM. After incubation, the mixture and reduced cytochrome c were added into the assay buffer, then subjected to measurement at 30 °C (Agilent Technologies, cary300). Slopes of OD550 for 1 min were calculated and corrected by a value of hpCcO.
FRET-Based Measurement of Mitochondrial ATP Concentration.
FRET-based measurement of mitochondrial ATP concentration in cardiomyocytes was measured as previously described (8, 33). Briefly, FRET signal was measured in cardiomyocytes infected with adenovirus encoding mit-AT1.03 with an Olympus IX-81 inverted fluorescence microscope (Olympus) using a PL APO 60×, 1.35 N.A., oil immersion objective lens (Olympus). Fluorescence emission from Mit-ATeam was imaged by using a dual cooled CCD camera (ORCA-D2; Hamamatsu Photonics) with a dichroic mirror (510 nm) and two emission filters (483/32 nm for CFP and 542/27 nm for YFP; A11400-03; Hamamatsu Photonics). Cells were illuminated using the CoolLED pE-1 excitation system (CoolLED) with a wavelength of 425 nm. Image analysis was performed using MetaMorph (Molecular Devices). The YFP/CFP emission ratio was calculated by dividing pixel by pixel (a YFP image with a CFP image after background subtraction).
Statistical Analyses.
The comparison between two groups was made by t test (two tailed). For MASC assay, comparison was made by repeated two-way ANOVA. A value of P < 0.05 was considered statistically significant. Data represent mean ± SEM.
Further methods are found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank T. Miyazaki (Cyclex) for making antibodies; Y. Okazaki and Y. Tokuzawa (Saitama Medical University) for measurement of CcO activity by cary300; Dr. Steven Coppen for critical reading of the manuscript; S. Ikezawa, E. Takada, and H. Shingu for technical assistance; M. Kobayashi, R. Maki, and the Center for Research Education in Osaka University for MS analysis; Y. Okada for secretarial support; and H. Shimada for discussion and advice. This research was supported by the Japan Society for the Promotion of Science through the “Funding Program for Next Generation World-Leading Researchers (NEXT Program),” initiated by the Council for Science and Technology Policy; grants-in-aid from the Ministry of Health, Labor, and Welfare-Japan; grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology-Japan; and grants-in-aid from the Japan Society for the Promotion of Science. This research was also supported by grants from Takeda Science Foundation, Japan Heart Foundation, Japan Cardiovascular Research Foundation, Japan Intractable Diseases Research Foundation, Japan Foundation of Applied Enzymology, Japan Medical Association, Uehara Memorial Foundation, Mochida Memorial Foundation, Banyu Foundation, Naito Foundation, Inoue Foundation for Science, Osaka Medical Research foundation for intractable diseases, Ichiro Kanehara Foundation, and Showa Houkoukai.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419767112/-/DCSupplemental.
References
- 1.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272(5265):1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 2.Morgan JE, Vakkasoglu AS, Lanyi JK, Gennis RB, Maeda A. Coordinating the structural rearrangements associated with unidirectional proton transfer in the bacteriorhodopsin photocycle induced by deprotonation of the proton-release group: A time-resolved difference FTIR spectroscopic study. Biochemistry. 2010;49(15):3273–3281. doi: 10.1021/bi901757y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoyama H, et al. A peroxide bridge between Fe and Cu ions in the O2 reduction site of fully oxidized cytochrome c oxidase could suppress the proton pump. Proc Natl Acad Sci USA. 2009;106(7):2165–2169. doi: 10.1073/pnas.0806391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogura T, Kitagawa T. Resonance Raman characterization of the P intermediate in the reaction of bovine cytochrome c oxidase. Biochim Biophys Acta. 2004;1655(1-3):290–297. doi: 10.1016/j.bbabio.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Yoshikawa S, et al. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280(5370):1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 6.Gospodarowicz D, Abraham JA, Schilling J. Isolation and characterization of a vascular endothelial cell mitogen produced by pituitary-derived folliculo stellate cells. Proc Natl Acad Sci USA. 1989;86(19):7311–7315. doi: 10.1073/pnas.86.19.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci USA. 1993;90(9):4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kioka H, et al. Evaluation of intramitochondrial ATP levels identifies G0/G1 switch gene 2 as a positive regulator of oxidative phosphorylation. Proc Natl Acad Sci USA. 2014;111(1):273–278. doi: 10.1073/pnas.1318547111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vukotic M, et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15(3):336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32(8):1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YC, et al. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15(3):348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukuda R, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129(1):111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 13.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsukihara T, et al. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995;269(5227):1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 15.Wilson DF, Gilmour MV. The low-temperature spectral properties of mammalian cytochrome oxidase. I. The enzyme in intact rat-liver mitochondria. Biochim Biophys Acta. 1967;143(1):52–61. doi: 10.1016/0005-2728(67)90108-9. [DOI] [PubMed] [Google Scholar]
- 16.Heibel GE, Anzenbacher P, Hildebrandt P, Schäfer G. Unusual heme structure in cytochrome aa3 from Sulfolobus acidocaldarius: A resonance Raman investigation. Biochemistry. 1993;32(40):10878–10884. doi: 10.1021/bi00091a043. [DOI] [PubMed] [Google Scholar]
- 17.Babcock GT, Callahan PM. Redox-linked hydrogen bond strength changes in cytochrome a: Implications for a cytochrome oxidase proton pump. Biochemistry. 1983;22(10):2314–2319. doi: 10.1021/bi00279a002. [DOI] [PubMed] [Google Scholar]
- 18.Fujikawa M, Yoshida M. A sensitive, simple assay of mitochondrial ATP synthesis of cultured mammalian cells suitable for high-throughput analysis. Biochem Biophys Res Commun. 2010;401(4):538–543. doi: 10.1016/j.bbrc.2010.09.089. [DOI] [PubMed] [Google Scholar]
- 19.Imamura H, et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci USA. 2009;106(37):15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muramoto K, et al. Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate. Proc Natl Acad Sci USA. 2010;107(17):7740–7745. doi: 10.1073/pnas.0910410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshikawa S, Tsukhihara T, Shinzawa-Itoh K. [Crystal structure of fully oxidized cytochrome c-oxidase from the bovine heart at 2.8 A resolution] Biokhimiia. 1996;61(11):1931–1940. [PubMed] [Google Scholar]
- 22.Tsukihara T, et al. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc Natl Acad Sci USA. 2003;100(26):15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klammt C, et al. Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat Methods. 2012;9(8):834–839. doi: 10.1038/nmeth.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denko N, et al. Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin Cancer Res. 2000;6(2):480–487. [PubMed] [Google Scholar]
- 26.Wang J, et al. Pancreatic beta cells lack a low glucose and O2-inducible mitochondrial protein that augments cell survival. Proc Natl Acad Sci USA. 2006;103(28):10636–10641. doi: 10.1073/pnas.0604194103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An HJ, et al. Higd-1a interacts with Opa1 and is required for the morphological and functional integrity of mitochondria. Proc Natl Acad Sci USA. 2013;110(32):13014–13019. doi: 10.1073/pnas.1307170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horan MP, Pichaud N, Ballard JW. Review: Quantifying mitochondrial dysfunction in complex diseases of aging. J Gerontol A Biol Sci Med Sci. 2012;67(10):1022–1035. doi: 10.1093/gerona/glr263. [DOI] [PubMed] [Google Scholar]
- 29.Saxena R, et al. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am J Hum Genet. 2006;79(1):54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 31.Diaz F. Cytochrome c oxidase deficiency: Patients and animal models. Biochim Biophys Acta. 2010;1802(1):100–110. doi: 10.1016/j.bbadis.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161(1):1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 33.Shintani Y, et al. Toll-like receptor 9 protects non-immune cells from stress by modulating mitochondrial ATP synthesis through the inhibition of SERCA2. EMBO Rep. 2014;15(4):438–445. doi: 10.1002/embr.201337945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.