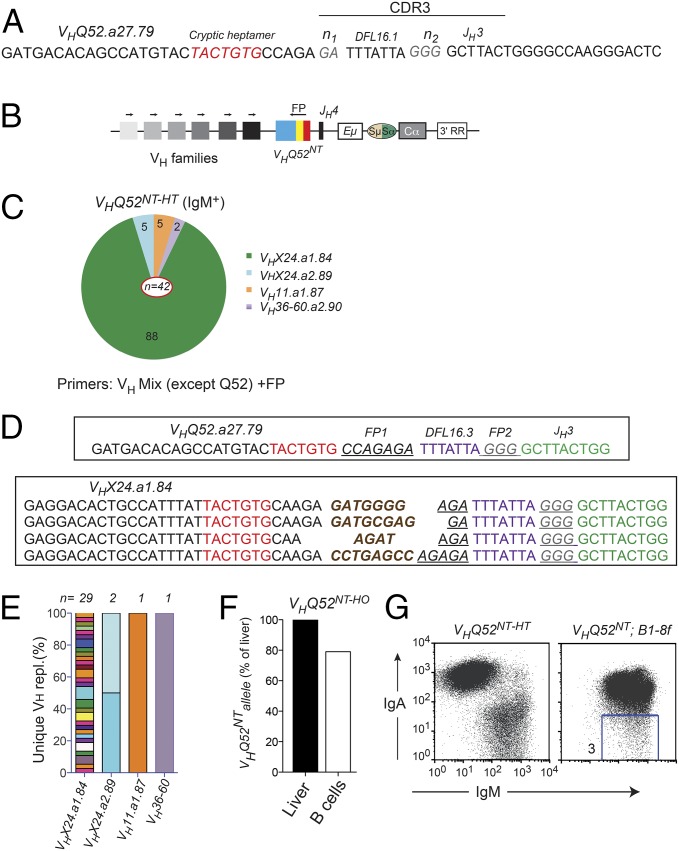
Fig. 4.
VHQ52NT is edited through VH replacement. (A) CDR3 nucleotide sequence of VHQ52NT. Cryptic heptamer and n-nucleotides are indicated, respectively, in red and gray. (B) PCR strategy to identify VH replacements. Arrows indicate PCR primers. Colored blocks symbolize VH families. An oligonucleotide annealing to a unique sequence (FP) within the CDR3 of VHQ52NT was used as a reverse primer. (C) Pie representation of VH gene use in VH replacements cloned from splenic IgM+ B cells of VHQ52NT-HT mice (n = 4). Numbers indicate frequency of rearrangements carrying the indicated VH genes. PCR primers used for the analysis are indicated. (D) CDR3 nucleotide sequence of representative VH replacements cloned from IgM+ B cells of VHQ52NT-HT mice using the VHX24.a1.84 donor VH gene. First line shows CDR3 sequence of the original VHQ52NT rearrangement. Underlined are VHQ52NT FP sequences indicated, respectively, as FP1 and FP2. De novo added n-nucleotides are labeled in brown. cRSSs are indicated in red. (E) Frequency of unique VH replacements cloned from IgM+ splenic B cells of VHQ52NT-HT mice, as assessed through CDR3 sequence analysis. Replacements using the same VH germ-line gene were grouped. (F) Southern blotting quantification of VHQ52NT gene copy number in splenic IgA+ B cells of VHQ52NT homozygous (HO) mice. Data were normalized for DNA input and represented as relative to VHQ52NT copy number in liver cells of transnuclear mice. (G) Representative flow cytometric analysis of splenic CD19+ B cells of VHQ52NT;B1-8f double IgH insertion mice (n = 4). Number within dot plot refers to frequency of IgM-only B cells.