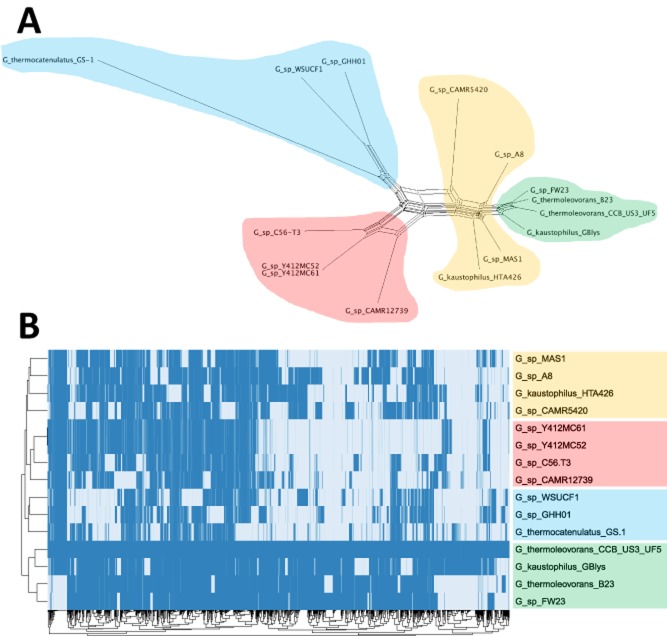
Fig 3.
Relationships among sequenced genomes within the G. kaustophilus clade resolved using whole-genome sequence data. The phylogenetic network in panel A was based on a concatenation of 1722 variant single-nucleotide sites in 1 874 967 nucleotides of the core genome present in all 15 genomes. The network was generated using the neighbornetalgorithm (Bryant and Moulton, 2004) implemented in the splitstree software package (Huson, 1998). The heat-map in B indicates the presence (dark blue) and absence (light blue) of each of 931 non-core genes from the genome of G. thermoleovorans CCB US3 UF6 across the same 15 genomes appearing in A. The gene-content clusters are shaded in the same colours in both panels. The heat-map was rendered using Raivo Kolde's pheatmap package in R (R Development Core Team, R, 2013).