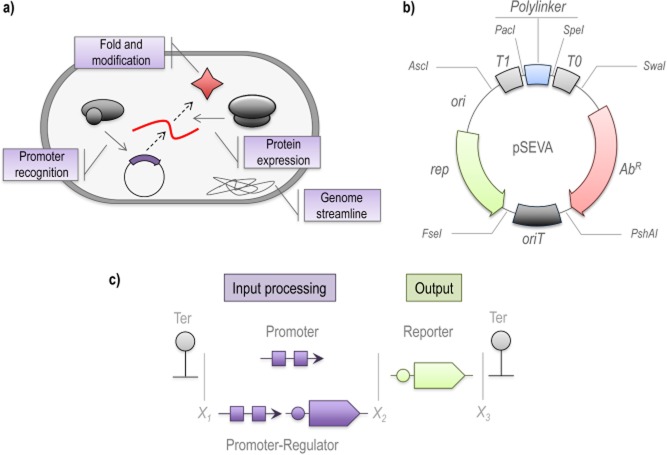
Fig 2.
Synthetic Biology may overcome limiting steps in activity-based metagenomic library screening. Current bottlenecks in functional metagenomics are related to (A) limitations in the host capabilities, (B) the performance of the genetic tools and (C) the availability of efficient screening methods.A. In the case of the host, critical steps related to the recognition of transcriptional and translational signals, as well as the folding and modification of the expressed enzyme need to be enhanced. Host performance might be improved by reducing the metabolic burden related to the expression of unnecessary genes.B. The use of semi-synthetic, high-efficiency genetic tools is essential for the construction of metagenomic libraries that can be maintained and screened in a wide number of microorganisms. The example shows the pSEVA bacterial vector, which is endowed with several functional features such as terminators, origin for transfer and an extensive polylinker optimized for use in several bacterial hosts.C. Genetic circuits constructed by combining input modules (e.g. promoters and regulators) and output devices (such as reporter proteins) assembled with a standard format that uses the same sets of restriction enzymes (represented by X1, X2, etc.). Such circuits facilitate the screening of enzymatic activities in metagenomic libraries. The standardization of the assembly process facilitates the combination of several independent modules to construct sophisticated activity-trigged biosensors.