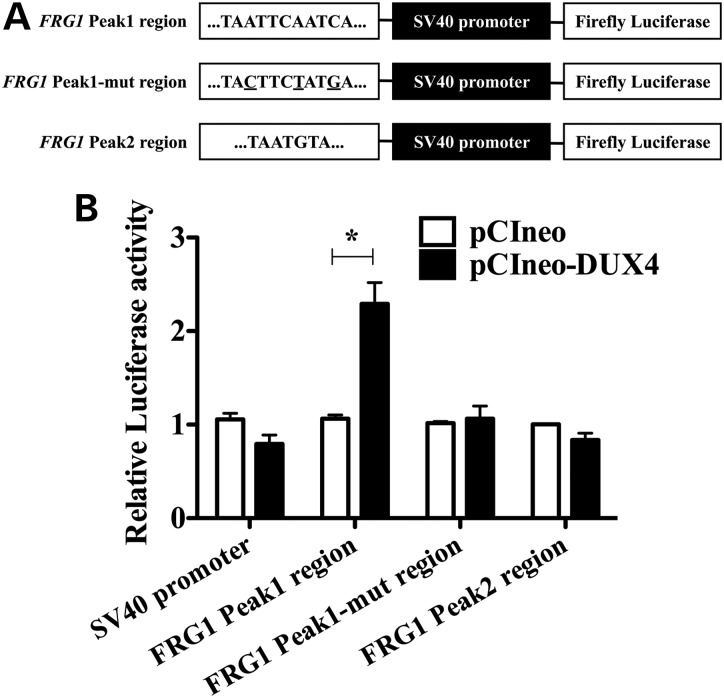
Figure 3.
FRG1 Peak1 region is sufficient for specific DUX4 transactivation. (A) Schematic representation of the constructs employed in luciferase assays. Genomic fragments of the FRG1 gene containing DUX4 putative binding sites were cloned upstream of the SV40 promoter, which controlled the expression of the Luciferase reporter gene. DUX4 core binding motifs within FRG1 genomic regions are reported. The three nucleotides of FRG1 Peak1 region that were mutated to generate FRG1 Peak1-mutated region are underlined. (B) FRG1 Peak1 region is able to transactivate the Luciferase reporter gene in the presence of DUX4, while the mutation in key DUX4 consensus sequence nucleotides abolishes the transactivation. DUX4 is not able to transactivate the Luciferase gene through FRG1 Peak2 region (paired t-test, *P < 0.05, n = 3, mean ± SEM).