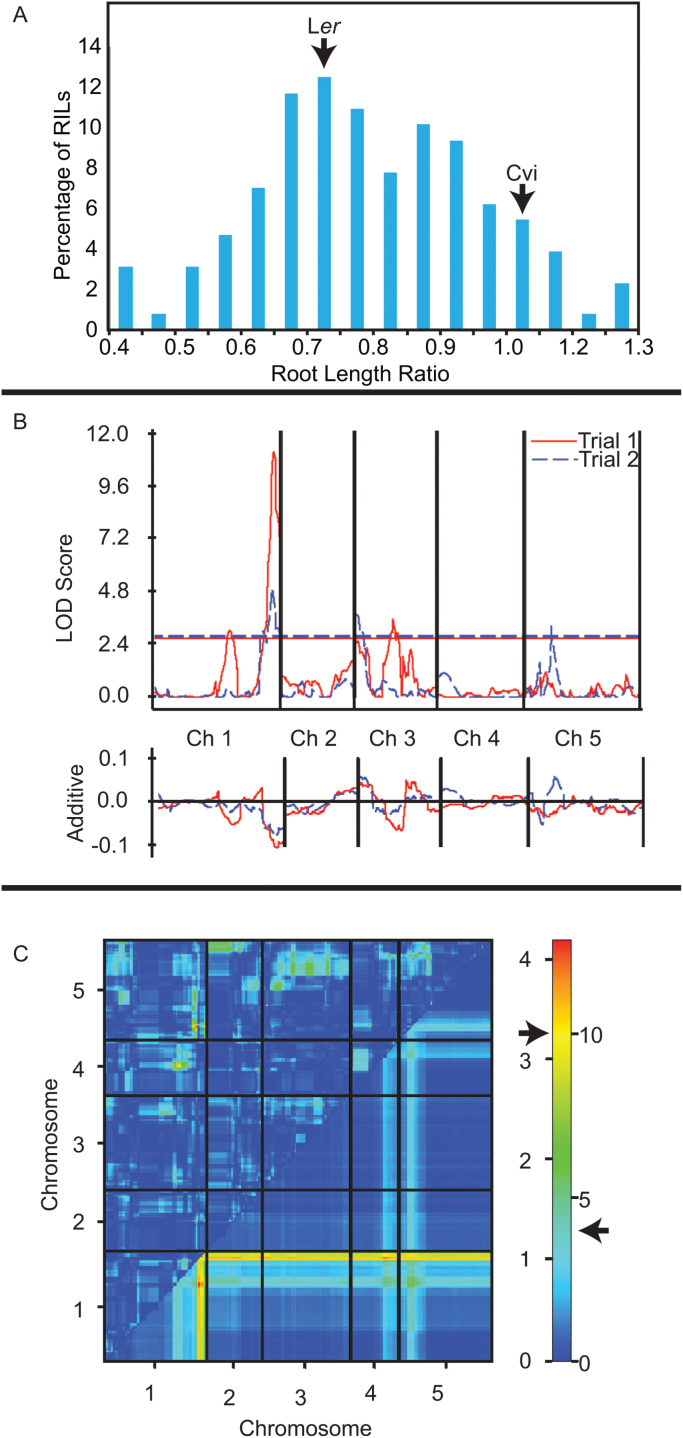
Fig. 2.
QTL analysis of root length responses to cadaverine in the Cvi/Ler RIL population. (A) Histogram showing root length ratios for the RILs on 100 μM cadaverine compared with control medium. (B) Composite interval mapping analysis for the root length ratio. The top portion of the graph gives the LOD score across each chromosome. The LOD significance threshold is shown as a horizontal line across the graph. The bottom graph shows the additive value toward the phenotype of each genomic region with respect to the Ler allele. (C) Two-dimensional QTL scan for the root length ratio from trial 1. The x- and y-axes represent positions along each chromosome. The region of the plot below the diagonal gives the additive QTL model, while the region above the diagonal shows epistatic interactions. A scale relating colour to LOD score is provided on the right of the heat map. Numbers on the left of this scale represent LOD scores for the epistatic portion of the plot, whereas those on the right represent scores for the additive model. The arrows indicate the significance thresholds for the epistatic (left) and additive (right) portions of the plot.