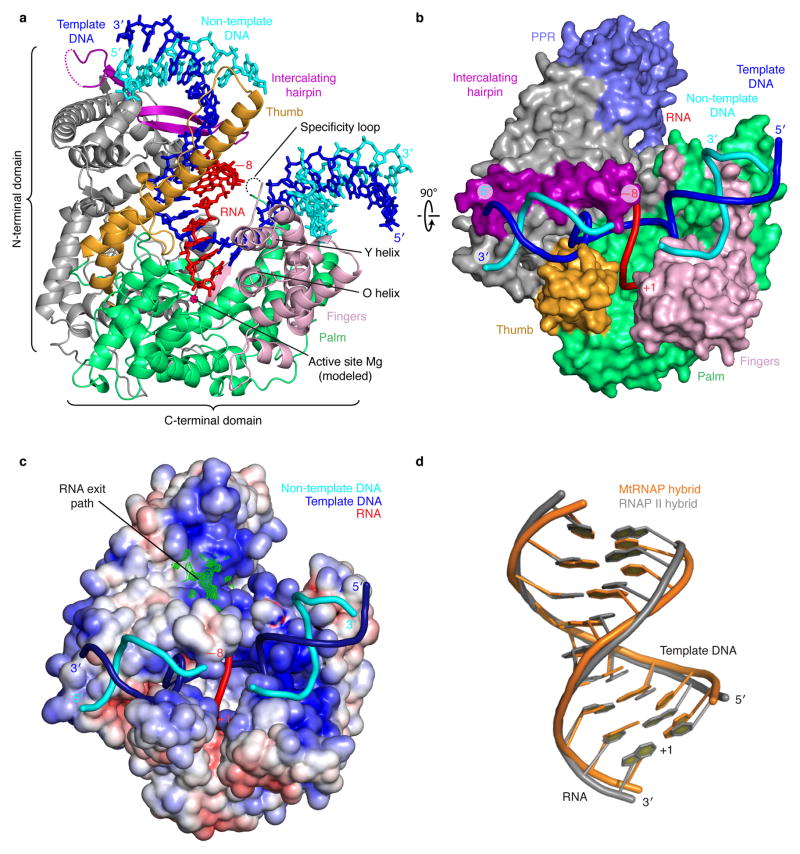
Figure 2. Structure of mtRNAP elongation complex determined by x-ray crystallography.
(a) Overview with mtRNAP depicted as a ribbon (thumb, orange; palm, green; fingers, pink; intercalating hairpin, purple), and nucleic acids as sticks (color code as in Fig. 1). A Mg2+ ion (magenta) was placed according to a T7 RNAP structure9. PPR domain was omitted for clarity.
(b) View of the structure rotated by 90° around a horizontal axis. The polymerase is depicted as a surface model and includes the PPR domain (slate). Nucleic acids are depicted as ribbons.
(c) Electrostatic surface representation of the mtRNAP elongation complex with template DNA (blue), non-template DNA (cyan) and RNA (red). The Fo-Fc electron density of the mobile 5′-RNA tail is shown as a green mesh (contoured at 2.5 σ).
(d) Superimposition of RNA–DNA hybrids in elongation complexes of mtRNAP (orange) and RNAP II (PDB 1I6H33, grey).