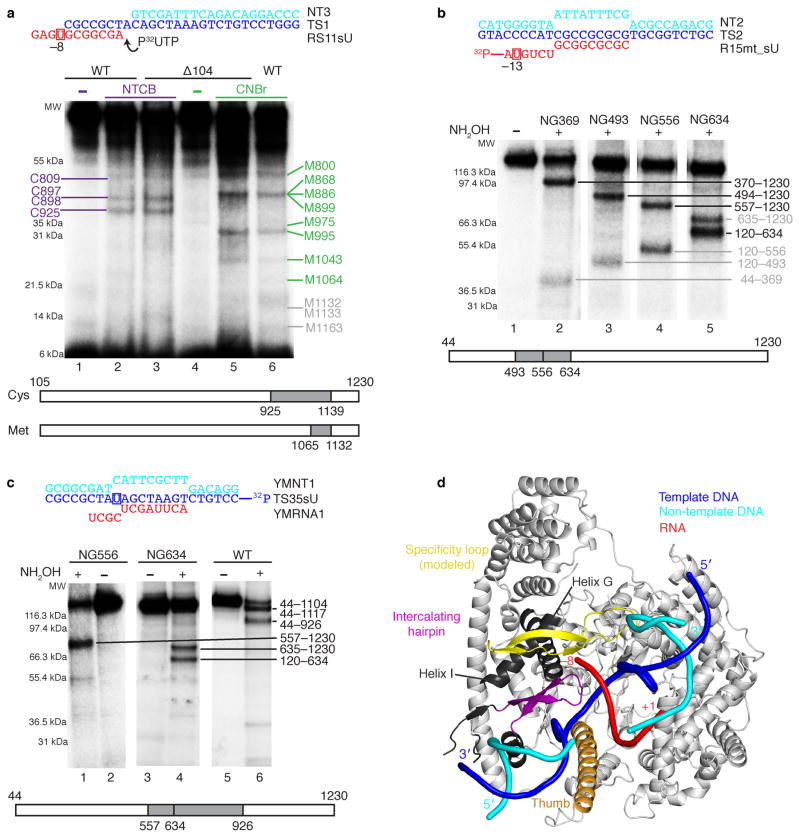
Figure 4. Analysis of mtRNAP–nucleic acid contacts by cross-linking experiments (see also Supplementary Fig. 5b and c online).
(a) RNA nucleotide –9 cross-links to the specificity loop of mtRNAP. The cross-linked complexes were treated with 2-nitro-5-thiocyano-benzoic acid (NTCB, lanes 2 and 3) or cyanogen bromide (CNBr, lanes 5 and 6). Positions of the cysteine (cys) and methionine (met) residues that produced labeled peptides are indicated in purple and green, correspondingly. Grey numbers indicate methionine residues that did not produce labeled peptides and the expected migration of these peptides.
(b) Mapping of the RNA–mtRNAP cross-link at RNA nucleotide –13 with different mtRNAP variants having a single hydroxylamin clevage site (NG) at a defined position. The cross-links were treated with hydroxylamine (NH2OH). The major cross-linked peptides are highlighted in black, minor (less than 10%) cross-linking sites in grey.
(c) Mapping of the template strand DNA–mtRNAP cross-link at nucleotide –8. The cross-links were treated with NH2OH as described above.
(d) Location of the cross-linked regions in mtRNAP elongation complex.
The T7 RNAP specificity loop was built into the mRNAP structure by homology modeling. The structural elements that belong to the identified cross-linked regions and lie within 3–5 Å from the photo cross-linking probe include the modeled specificity loop (yellow, residues 1080–1108), part of the thumb (orange, residues 752–791) and part of the intercalating hairpin (purple, residues 605–623). Cross-linked regions that are not part of a defined structural element are shown in dark grey (e.g. helix G residues 587–571 and helix I residues 570–586).