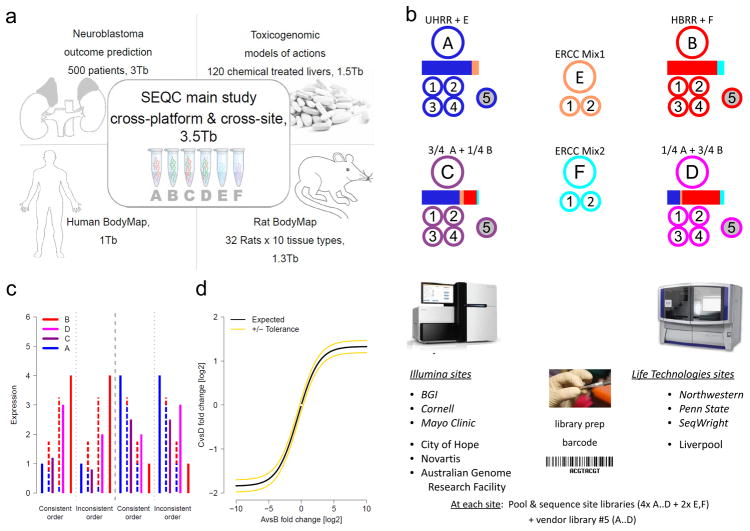
Figure 1.
The SEQC (MAQC-III) project and experimental design. (a) Overview of projects. We report on a group of studies assessing different sequencing platforms in real-world use cases, including transcriptome annotation and other research applications, as well as clinical settings. This paper focuses on the results of a multi-center experiment with built-in ground truths. (b) Main study design. Similar to the MAQC-I benchmarks, we analysed RNA samples A to D: Samples C and D were created by mixing the well-characterized samples A and B in 3:1 and 1:3 ratios, respectively. This allows tests for titration consistency (c) and the correct recovery of the known mixing ratios (d). Synthetic RNAs from the External RNA Control Consortium (ERCC) were both pre-added to samples A and B before mixing and also sequenced separately to assess dynamic range (samples E and F). Samples were distributed to independent sites for RNA-seq library construction and profiling by Illumina’s HiSeq 2000 (3 official + 3 inofficial sites) and Life Technologies’ SOLiD 5500 (3 official sites + 1 inofficial site). Unless mentioned otherwise, data presented shows results from the three official sites (italics). In addition to the four replicate libraries each for samples A to D per site, for each platform, one vendor-prepared library A5…D5 was being sequenced at the official sites, giving a total of 120 libraries. At each site, every library has a unique bar-code sequence and all libraries were pooled before sequencing, so each lane was sequencing the same material, allowing a study of lane specific effects. To support a later assessment of gene models, we sequenced samples A and B by Roche 454 (3x, no replicates, see Supplementary Notes Section 2.5). (c) Schema illustrating tests for titration order consistency. Four examples are shown. The dashed lines represent the ideal mixture of samples A and B (blue and red) expected for samples D and C (magenta and dark purple). (d) Schema illustrating a consistency test for recovering the expected sample mixing ratio. The yellow lines mark a 10% deviation from the expected response (black) for a perfect mixing ratio. Both tests (c) and (d) will reflect both systemic distortions (bias) and random variation (noise).