Deficiency of the ubiquitin-editing enzyme A20 in the hematopoietic system causes anemia, lymphopenia, and postnatal lethality as the result of defective hematopoietic stem cell function.
Abstract
A balance between quiescence and proliferation is critical for proper maintenance of the hematopoietic stem cell (HSC) pool. Although a lot is known about hematopoiesis, molecular mechanisms that control HSC quiescence remain largely unknown. The ubiquitin-editing enzyme A20 functions as a central regulator of inflammation and adaptive immunity. Here, we show that a deficiency of A20 in the hematopoietic system causes anemia, lymphopenia, and postnatal lethality. Lack of A20 in HSCs results in diminished pool size, impaired radioprotection, defective repopulation, and loss of quiescence. A20-deficient HSCs display increased IFN-γ signaling, caused by augmented NF-κB activation. Strikingly, deletion of both IFN-γ and A20 in hematopoietic cells results in partial rescue of the HSC phenotype. We anticipate that our experiments will facilitate the understanding of mechanisms through which A20-mediated inflammatory signals control HSC quiescence and functions.
Hematopoiesis is a well-orchestrated process through which the most primitive cells of the hematopoietic system, i.e., hematopoietic stem cells (HSCs), differentiate into mature cells of the myeloid-erythroid and lymphoid lineages (Morrison et al., 1995; Orford and Scadden, 2008; Orkin and Zon, 2008). HSCs are rare and reside in specialized niches located within the BM. In the absence of specific mitogenic stimuli, HSCs remain in a quiescent or dormant state (Trumpp et al., 2010). However, under stress conditions, such as bleeding, myeloablation, total body irradiation, and infection, HSCs can enter into an active proliferative state (Passegué et al., 2005). Although HSCs are controlled mainly by intrinsic pathways, often HSCs need and respond to external stimuli such as cytokines, chemokines, and cell–cell contacts. Whenever HSCs divide into daughter cells, the fates of the daughter cells, including life versus death and self-renewal versus differentiation, need to be tightly regulated because defects in these cell fate decisions will have detrimental consequences, including BM failure and hematologic malignancies (Passegué et al., 2005).
Self-renewal and differentiation of HSCs are complex processes and are dependent on the immediate “turning on” or “turning off” of various cytokine receptors, signal transducers, transcription factors, and cell cycle inhibitors. Although transcriptional regulation of gene expression and the involvement of transcription factors in hematopoiesis have been studied to a greater extent, the role of posttranslational modifications (PTMs) of proteins, in particular ubiquitination, in the regulation of hematopoiesis remains largely unknown. Recent studies, including our own, have highlighted the importance of the ubiquitin proteasome system in the development and functions of normal HSCs and leukemic stem cells (Rathinam et al., 2008, 2010, 2011; Rathinam and Flavell, 2010; Moran-Crusio et al., 2012). Even though these studies have provided clues regarding the physiological relevance of PTMs in hematopoiesis, a clearer understanding of the significance of PTMs mediated by the ubiquitin proteasome system in early hematopoiesis remains elusive. Moreover, these studies were based on the functions of E3 ubiquitin ligases, such as c-Cbl, Itch, and Fbxw7, and the role of deubiquitinases (DUBs) in early hematopoiesis and in stem cell biology needs to be explored.
A20 (also known as Tnfaip3 and referred as A20 henceforth) is a potent antiinflammatory signaling molecule that restricts multiple intracellular signaling cascades (Ma and Malynn, 2012). A20 is an ∼90-kD protein that belongs to the ovarian tumor (OTU) family of DUBs. A unique feature of A20 is that it contains an N-terminal cysteine protease/DUB domain (which is necessary for the deubiquitylating functions) and a C-terminal zinc finger (ZNF) domain (which confers the E3 ubiquitin ligase functions). Thus, A20 can be classified as a dual-function ubiquitin-editing enzyme (Wertz et al., 2004). A20 catalyzes the K48-linked ubiquitylation of target proteins through its C-terminal ZNF domain, an action which directs the target proteins for proteasomal degradation. Concurrently, A20 removes K63-linked ubiquitin chains from its target proteins (through its DUB activity), which not only inactivates the signaling function of the targets, but might also facilitate its K48-linked ubiquitylation and degradation (Wertz et al., 2004). The negative signaling function of A20 involves deconjugation of K63-linked ubiquitin chains from TRAF6 and RIP1, which are central players of the TLR and TNF receptor (TNFR) pathways (Sun, 2008). In addition, A20 also mediates deubiquitylation of RIP2 and therefore acts as a negative regulator of NF-κB signaling (Hitotsumatsu et al., 2008; Sun, 2008; Vereecke et al., 2009; Hymowitz and Wertz, 2010).
In view of the fact that A20 has a key role in the control of inflammation and that inflammatory signals can impact HSC development and functions, we hypothesized that A20 functions as a critical regulator of the HSC pool. To validate this, we generated and investigated mice that lack A20 expression in HSCs (and in their progeny). Deficiency of A20 in the hematopoietic system resulted in reduced body size and weight and postnatal lethality. BM and thymic cellularity was remarkably reduced, whereas extramedullary hematopoiesis in the spleen and liver was increased in A20 mutant mice. Loss of A20 led to a striking reduction of the long-term HSC (LT-HSC) pool and an increase of the short-term HSC (ST-HSC) pool. A20-deficient HSCs exhibited compromised abilities to provide radioprotection and hematopoietic reconstitution in lethally irradiated WT recipients. Hematopoietic stem and progenitor cells (HSPCs) of A20 mutant mice showed hyperproliferation, loss of quiescence, and reduced expression of p57. Our mechanistic experiments in A20-deficient mice identified augmented IFN-γ signaling in HSCs and elevated IFN-γ expression by hematopoietic cells, as the result of increased NF-κB binding to the promoter of Ifn-γ. Notably, loss of IFN-γ in A20-deficient mice partially rescued the HSC phenotype. Overall, our experiments identified the importance of A20-mediated suppression of inflammatory signals in HSC homeostasis and highlighted ubiquitin editing as a novel mechanism in the control of HSC quiescence.
RESULTS
A20 deficiency in HSCs causes postnatal lethality
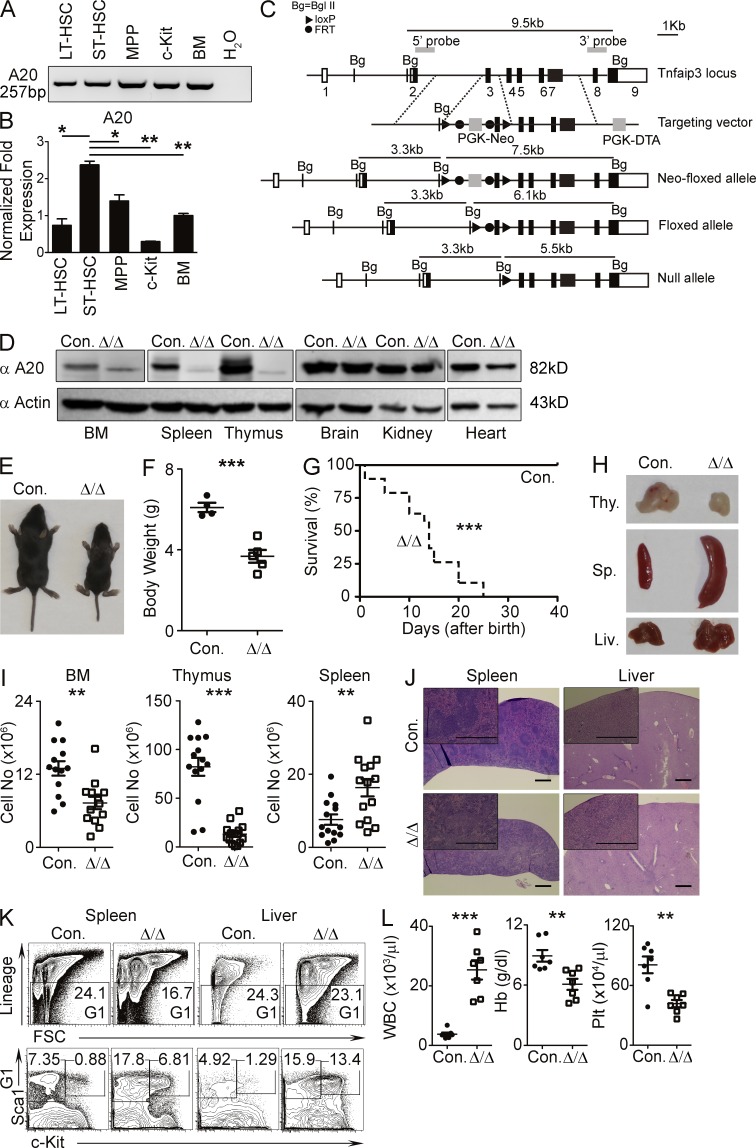
To identify whether A20 is expressed during early hematopoiesis, we divided BM Lineage−Sca1+c-Kit+ (LSK) cells from C57BL/6 WT mice into LT-HSCs (CD150+CD48−LSK), ST-HSCs (CD34+Flt3−LSK), and multipotent progenitors (MPPs; CD34+Flt3+LSK), sorted them individually, and quantified expression levels of A20 mRNA by RT-PCR. Indeed, A20 was expressed in all three LSK subsets (i.e., LT-HSCs, ST-HSCs, and MPPs), in c-Kit+Lin− myeloid progenitor cells, and in total BM cells (Fig. 1 A). Of note, the highest expression levels of A20 were found in ST-HSCs (Fig. 1 B). To study the functions of A20 in HSCs, we generated mice with loxP-flanked alleles of the gene encoding mouse A20 (Fig. 1 C). To analyze Cre-mediated ablation of A20 expression, we crossed A20 Floxed mice with the ubiquitous-Cγ1-Cre animals to obtain A20Flox/Flox Cγ1-Cre (designated as A20−/−) mice. To test whether A20 is deleted in these mice, mesenchymal embryonic fibroblasts (MEFs) were generated, from either control or A20−/− embryos, and stimulated with TNF (as it induces A20 expression in MEFs). Western blot analysis indicated the presence and induction of A20 protein in control MEFs, but it was completely abolished in A20−/− MEFs (not depicted). We also used antibodies that recognize the N-terminal epitope of A20, which may still present in the truncated A20 protein, and confirmed that no aberrant A20 protein was expressed in these cells (not depicted). To delete A20 in HSCs, we crossed A20F/F mice with the Vav-Cre transgenic mice (Ogilvy et al., 1999) to generate A20Flox/FloxVavCre/+ (called A20Hem-KO here) mice and confirmed that A20 protein was predominantly absent in the hematopoietic organs, such as BM, spleen, and thymus. As expected, A20 expression was intact in nonhematopoietic organs, such as brain, kidney, and heart (Fig. 1 D).
Figure 1.
A20 deficiency leads to pathological hematopoiesis. (A and B) Reverse transcription–PCR (A) and real-time PCR (B) analyses showing A20 mRNA expression in the indicated hematopoietic subsets of WT BM. Expression levels of A20 were normalized to Hprt levels. (C) Scheme representing the Tnfaip3 gene targeting strategy. (D) Western blot analysis of A20 protein in cells from the indicated organs of A20Hem-KO and WT control mice. (E) Representative images of control and A20Hem-KO mice showing body size. (F) Total body weight of control (n = 4) and A20Hem-KO (n = 5) mice at 14 d of birth. (G) Kaplan-Meier survival curve analysis of A20Hem-KO (n = 15) and control (n = 15) mice. Significance (***, P < 0.001) was assessed using the log-rank test. (H) Representative images of thymus, spleen, and liver of 14-d-old A20Hem-KO and control mice. (I) Cellularity of BM (two femurs and two tibias), thymus, and spleen of 14-d-old A20Hem-KO (n = 13–14) and control (n = 13–14) mice. Data are representative of 10 independent experiments. (J) Representative hematoxylin and eosin images (bars, 0.5 mm) of spleen and liver of 14-d-old A20Hem-KO and control mice. In each panel, a high-magnification image is shown at the top left corner. (A, B, D, and J) Data are representative of two independent experiments. (K) FACS plots of LSK and Lin−c-Kit+Sca1− cells in the spleen and liver of 14-d-old A20Hem-KO and control mice. Data are representative of three independent experiments. (L) Differential blood count analysis of 14-d-old A20Hem-KO (n = 7) and control (n = 7) mice. Data are pooled from seven independent experiments. All data represent mean ± SEM. Two-tailed Student’s t tests were used to assess statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
A20Hem-KO mice were born at Mendelian ratios; however, they exhibited ∼50% reduction of body size and weight (Fig. 1, E and F), as compared with their littermate controls. Strikingly, almost all A20Hem-KO mice died before their weaning age (∼21 d) and had a median lifespan of 14 d (Fig. 1 G). Analysis of 14-d-old A20Hem-KO mice indicated (Fig. 1 H) thymic atrophy (reduced thymus), splenomegaly (increased spleen size), and hepatomegaly (enlarged liver). Cell count analysis showed an ∼50% reduction in BM cellularity, ∼90% reduction in thymic cellularity, and ∼400% increase in splenic cellularity of A20Hem-KO mice (Fig. 1 I). Histopathological analysis of spleen and liver of A20Hem-KO mice revealed disrupted architecture, infiltration of leukocytes (Fig. 1 J), and extramedullary hematopoiesis (Fig. 1 K). Peripheral blood count analysis of A20Hem-KO mice indicated elevated WBC counts and reduced hemoglobin and platelets counts (Fig. 1 L). Collectively, these data suggest that A20 has an indispensable role in restricting pathological hematopoiesis.
A20 controls the HSC pool
To determine whether the HSC pool of A20Hem-KO mice was altered, we performed immunophenotyping experiments of HSCs and HSPCs by flow cytometry. Analysis of the LSK compartment, consisting of LT-HSCs, ST-HSCs, and MPPs, revealed a remarkable increase (3.2% in control and 17.9% in KO) of relative numbers in the BM of A20Hem-KO mice (Fig. 2 A). Further investigation of LSK cells indicated a striking reduction of LT-HSCs (4.8% in control and 0.09% in KO) and MPPs (64% in control and 8% in KO) in the BM of A20Hem-KO mice (Fig. 2 A). Interestingly, frequencies of ST-HSCs were increased (27% in control and 86% in KO) in the BM A20Hem-KO mice (Fig. 2 A). Next, we enumerated the absolute numbers of HSPC subsets of the BM. Even though the relative frequencies of LSK cells were increased in A20Hem-KO mice, the absolute numbers of LSK cells were comparable between controls and A20Hem-KO mice (Fig. 2 B). Consistent with their relative numbers, the absolute numbers of LT-HSCs and MPPs were decreased, and the absolute numbers of ST-HSCs were increased in the BM of A20Hem-KO mice (Fig. 2 B).
Figure 2.
The HSPC pool is attenuated in A20-deficient mice. (A) FACS plots of LSK cells, LT-HSCs, ST-HSCs, and MPPs from the BM (two femurs and two tibias) of 14-d-old A20Hem-KO and control mice. Data are representative of 10 independent experiments. (B) Absolute numbers of LSK cells, LT-HSCs, ST-HSCs, and MPPs from the BM (two femurs and two tibias) of 14-d-old A20Hem-KO (n = 5) and control (n = 5) mice. (C) FACS plots of SP cells in the BM of 14-d-old A20Hem-KO and control mice. Data are representative of three independent experiments. (D) Relative frequencies of SP cells in the BM of 14-d-old A20Hem-KO (n = 3) and control (n = 3) mice. Data are pooled from three independent experiments. (E) FACS plots of quiescent HSCs and MPP subsets in the BM of 14-d-old A20Hem-KO (n = 4) and control (n = 4) mice. Data are representative of eight independent experiments. All data represent mean ± SEM. Two-tailed Student’s t tests were used to assess statistical significance (*, P < 0.05; ***, P < 0.001).
To independently confirm these findings through an alternative technique, we performed “side population” (SP) analysis (Goodell et al., 1996). This technique utilizes the fluorescent DNA-binding dye Hoechst 33342 to identify and purify mouse HSCs in the BM, as only HSCs, but not differentiated hematopoietic cells, will have the abilities to pump out the Hoechst 33342 dye. Our SP analysis identified a remarkable decrease (0.047% in control and 0.015% in KO) of SP cells in the BM of A20Hem-KO mice (Fig. 2, C and D).
To further strengthen these data, we adopted the immunophenotyping strategy, developed by the Trumpp laboratory (Wilson et al., 2008), that distinguishes HSPCs of the mouse BM into five distinct stages: HSC (LSK CD150+CD48−CD34−Flt3−), MPP1 (LSK CD150+CD48−CD34+Flt3−), MPP2 (LSK CD150+CD48+CD34+Flt3−), MPP3 (LSK CD150−CD48+CD34+Flt3−), and MPP4 (LSK CD150−CD48+CD34+Flt3+). According to this study (Wilson et al., 2008), the most primitive HSC (LSK CD150+CD48−CD34−Flt3−) fraction remains largely quiescent, and the transition from MPP1 to MPP4 stage is accompanied by a gradual loss of quiescence and an increase of proliferation. As expected, we detected almost no HSCs (13% in control and 0% in KO), slightly increased MPP1 (82% in control and 100% in KO), modestly reduced MPP2 (92% in control and 63% in KO), remarkably increased MMP3 (26% in control and 89% in KO), and highly reduced MPP4 (66% in control and 6% in KO) in the BM of A20Hem-KO mice (Fig. 2 E). These data suggested that the more quiescent HSC fraction was completely depleted and the transition from MPP3 to MPP4 was severely affected in the absence of A20-mediated functions in HSCs. Overall, the immunophenotyping experiments demonstrated that the HSC pool is remarkably reduced in the BM of A20Hem-KO mice.
A20-deficient HSCs have compromised functions
To assess the functions of A20 mutant HSCs, we performed BM transplantation (BMT) experiments. Transfer of A20-deficient BM cells (106) into lethally (10 Gy) irradiated WT animals failed to provide radioprotection in ∼70% of the recipients (Fig. 3 A). Even though the remaining ∼30% of recipients (which received A20-deficient BM) survived for >3 wk, they developed severe leukopenia, anemia, and thrombocytopenia, starting at 2 wk after transplantation (Fig. 3 B). In these survivors, A20 mutant–derived chimerism was >95% at 2 wk after transplantation, but it was dropped to ∼10% at 4 wk after transplantation (Fig. 3, C and D). Transfer of BM from the survivors that showed a donor chimera of ∼10% into lethally irradiated WT secondary recipients failed to provide detectable levels (<1%) of donor chimerism at 4 wk after transplantation (Fig. 3 E). We repeated these experiments using purified LSK cells and obtained similar results (not depicted). Based on the fact that we detected A20 mutant–derived hematopoiesis at 2 wk after transplantation, we reasoned that homing defects might not be responsible for the reduced engraftment in recipients. To substantiate this, we performed homing experiments using CFSE-labeled LSK cells and found that the homing capacities were comparable between control and A20Hem-KO HSPCs (Fig. 3 F).
Figure 3.
Loss of A20 causes impaired HSC functions. (A) Kaplan-Meier survival curve analysis of WT recipients (CD45.1+) that received BM of either A20Hem-KO or control mice (CD45.2+) after lethal (10 Gy) irradiation. Each group represents data from a pool of 13 animals. Significance (**, P < 0.004) was assessed using the log-rank test. (B) Differential blood count analysis of WT congenic (CD45.1+) recipients that received BM cells from either A20Hem-KO or WT mice, at 2 wk after transplantation. Data are pooled from three mice per group. (C) FACS plots of donor (either WT or A20Hem-KO)-derived chimera (CD45.2+) in the peripheral blood of WT congenic (CD45.1+) recipients, at 2 wk after transplantation. (D) Frequencies of donor (CD45.2+)-derived hematopoiesis, at 4 wk after transplantation, in the peripheral blood of WT (CD45.1+) recipients that survived lethal irradiation (survivors of A). Data are pooled from five mice in the control group and four mice in the A20Hem-KO group. (E) Serial transplantation experiments: Frequencies of donor (CD45.2+) cells in the peripheral blood of secondary WT recipients, at 4 wk after transplantation. Data are pooled from five mice per group. (F) FACS plots indicating frequencies of CFSE+ cells in the BM of lethally irradiated WT recipients that received CFSE-labeled LSK cells of either A20Hem-KO or WT control mice at 20 h after injection. Data are representative of two independent experiments with four mice per group. (G and H) Frequencies of donor (CD45.2+) hematopoiesis in the peripheral blood (G) and BM, spleen, and thymus (H) of WT recipients, at 4 wk after transplantation. Recipients received mixed chimera containing donor (CD45.2+) BM cells (from either A20Hem-KO or control mice) and WT competitor (CD45.1+) BM cells at a ratio of 1:1 (G) and 3:1 (G and H), respectively. Data are representative of two independent experiments with four mice per group (G) and four mice for WT and five mice for A20Hem-KO groups (H). (I and J) Frequencies of donor (CD45.2+)-derived cells in total CD11b+ (I) and CD19+ (J) fractions of the peripheral blood from WT recipients, at 4 wk after transplantation. Recipients received mixed chimera containing donor (CD45.2+) BM cells (from either A20Hem-KO or control mice) and WT competitor (CD45.1+) BM cells at a ratio of 1:1 and 3:1, respectively. Data are representative of two independent experiments with four mice per group. (K) Frequencies of donor (CD45.2+)-derived cells in the BM, spleen, thymus, and peripheral blood of WT recipients, at 2 and 9 wk after transplantation. Recipients received mixed chimera containing donor (CD45.2+) BM cells (from either A20Hem-KO or control mice) and WT competitor (CD45.1+) BM cells at a ratio of 5:1, respectively. (L) Frequencies of donor (CD45.2+)-derived cells in total CD11b+ and CD19+ fractions of the peripheral blood from WT recipients, at 2 and 9 wk after transplantation. Recipients received mixed chimera containing donor (CD45.2+) BM cells (from either A20Hem-KO or control mice) and WT competitor (CD45.1+) BM cells at a ratio of 5:1, respectively. (K and L) Data are representative of two independent experiments with three mice per group. All data represent mean ± SEM. Two-tailed Student’s t tests were used to assess statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Next, we tested the potential of A20-deficient BM to repopulate the hematopoietic system under competitive conditions. A20-deficient and control BM cells (CD45.2+) were mixed with congenic (CD45.1+) WT BM cells at a ratio of either 1:1 or 3:1 (donor/competitor) and were injected into lethally irradiated congenic (CD45.1+) recipients. Analysis of donor-derived chimera at 4 wk after transplantation indicated a significant reduction of donor-derived (CD45.2+) hematopoiesis (Fig. 3, G and H), as well as multilineage differentiation (Fig. 3, I and J), in recipients that received A20 mutant BM. Because no detectable levels of A20 mutant–derived chimerism were seen in recipients at 4 wk after transplantation, we increased the donor/competitor ratio to 5:1 and injected into lethally irradiated WT recipients. Analysis of donor-derived chimera indicated that even though A20 mutant–derived hematopoiesis was detectable (albeit ∼50% lower than that of the control group) in recipients at 2 wk after transplantation, it was undetectable in the BM, spleen, thymus, and peripheral blood at 9 wk after transplantation (Fig. 3, K and L). Collectively, these data indicate that A20 deficiency impairs HSC functions.
A20 regulates HSC quiescence
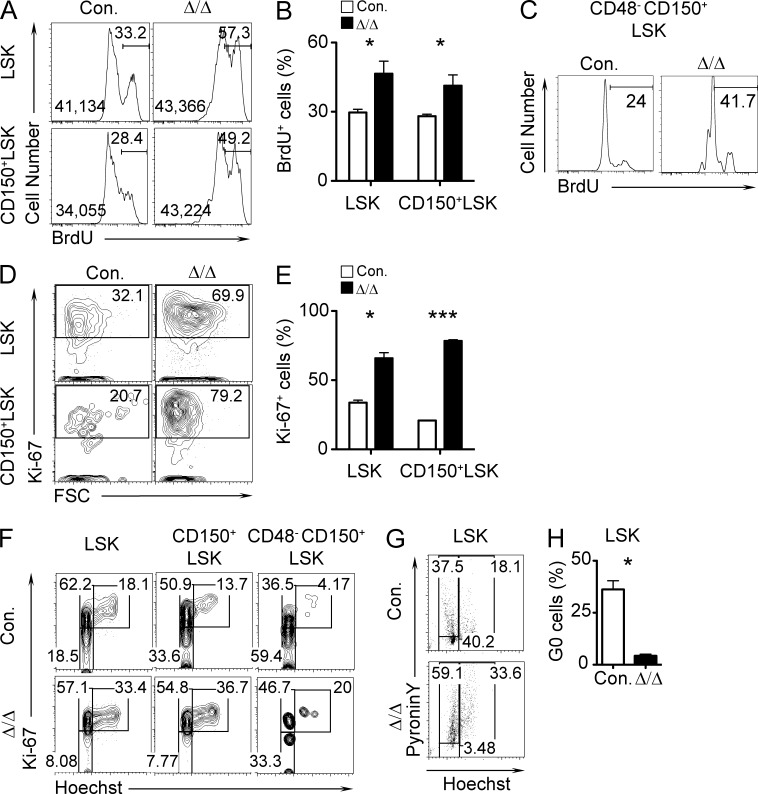
Our analysis of A20Hem-KO mice indicated a striking reduction of LT-HSCs in the BM (Fig. 2 A). Loss of LT-HSCs in A20 mutant BM could be explained by either increased apoptosis of LT-HSCs or increased proliferation in conjunction with decreased self-renewal of LT-HSCs. To identify the reason or reasons for the loss of the LT-HSC pool in A20 mutant mice, we first performed apoptosis experiments using Annexin V and 7-AAD. Data of these experiments indicated comparable frequencies of apoptosis between control and A20Hem-KO mice (not depicted). These data were further validated using TUNEL assays, and we obtained consistent results (not depicted). Next, we performed proliferation experiments using BrdU (Hock et al., 2004). Data of these experiments indicated that the frequencies of BrdU+ cells and that the intensities of incorporated BrdU were augmented both in total LSK and in CD150+LSK cells from 14-d-old A20Hem-KO mice (Fig. 4, A and B). Because LT-HSCs were almost absent in 14-d-old A20Hem-KO mice, we repeated BrdU assays with 2-d-old (P2) pups and observed that A20 mutant CD48−CD150+LSK cells proliferated more rapidly than control cells (Fig. 4 C). Consistent with this, Ki-67 expression levels were also increased in A20 mutant LSK and CD150+LSK cells (Fig. 4, D and E). Next, we measured HSC quiescence using Hoechst and Ki-67. As expected, the frequencies of LSK, CD150+LSK, and LT-HSC fractions at G0 phase were reduced in the BM of A20Hem-KO mice (Fig. 4 F). Finally, we performed Pyronin Y staining to distinguish quiescent HSPCs from proliferating HSPCs (Cheng et al., 2000). Pyronin Y analysis of A20 mutant BM revealed a striking reduction (40% in control and 3.5% in KO) of LSK cells in the G0 (Pyronin Y−Hoechst−) phase of the cell cycle (Fig. 4, G and H). Collectively, these data indicated that A20 deficiency might cause loss of HSC quiescence and premature depletion of the HSC pool.
Figure 4.
Deficiency of A20 results in impaired HSC quiescence. (A and B) Representative FACS plots (A) and bar graphs (B) indicating frequencies of BrdU+ cells in LSK and CD150+LSK fractions in the BM of 14-d-old A20Hem-KO and control mice, after 16-h pulse with BrdU. (A) In each histogram, the median fluorescence intensities (left) and percentages of cells under the gate (right) are indicated. Data are representative of two independent experiments with three mice per group. (C) FACS plots indicating frequencies of BrdU+ LT-HSCs from the livers of P2 pups after 16-h pulse with BrdU. Data are representative of two independent experiments with two pups per group. (D and E) Representative FACS plots (D) and cumulative frequencies (E) of Ki-67+LSK and CD150+LSK cells from 14-d-old A20Hem-KO and control mice. (F) FACS plots indicating frequencies of HSPCs in G0 (Ki-67− and Hoechst−), G1 (Ki-67+ and Hoechst−), and G2/S (Ki-67+ and Hoechst+) phase of cell cycle in the BM of 14-d-old A20Hem-KO and control mice. (G) Representative FACS plots indicating frequencies of HSPCs in G0 (Pyronin Y− and Hoechst−), G1 (Pyronin Y+ and Hoechst−), and G2/S (Pyronin Y+ and Hoechst+) phases of cell cycle in the BM of 14-d-old A20Hem-KO and control mice. (H) Bar graph showing the frequencies of HSPCs in the G0 (Pyronin Y− and Hoechst−) phase of the cell cycle in the BM of 14-d-old A20Hem-KO and control mice. (D, E, G, and H) Data are representative of two independent experiments with two mice per group. All data represent mean ± SEM. Two-tailed Student’s t tests were used to assess statistical significance (*, P < 0.05; ***, P < 0.001).
A20-deficient hematopoietic cells show increased NF-κB signaling
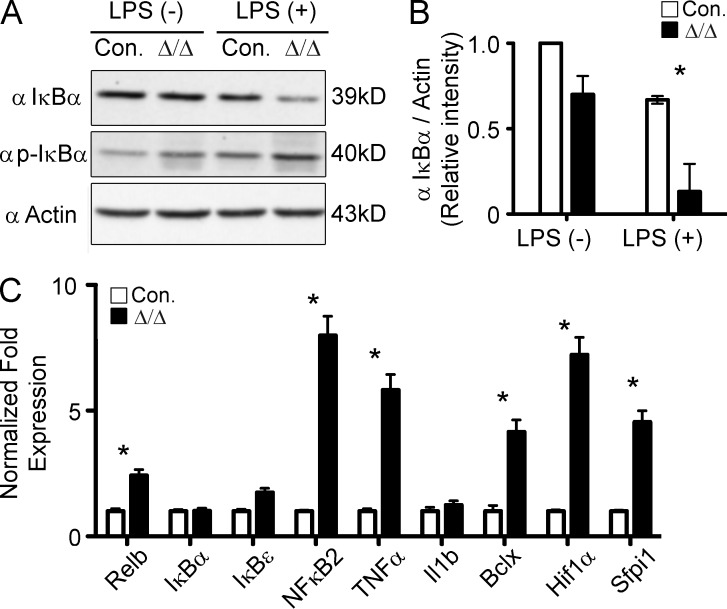
A20 is a key negative regulator of NF-κB signaling, and loss of A20 results in augmented NF-κB signal transduction (Vereecke et al., 2009; Hymowitz and Wertz, 2010; Ma and Malynn, 2012). To test whether NF-κB signaling was increased in A20Hem-KO mice, we measured IκBα expression and activation. Hematopoietic cells from control and A20 mutant mice were stimulated in vitro with LPS and assessed for protein expression. Western blot data indicated a baseline reduction of total IκBα protein in unstimulated cells of A20 mutant mice. However, in response to LPS, total IκBα levels were strikingly reduced and phospho- IκBα protein levels were increased in the hematopoietic cells of A20 mutant mice (Fig. 5, A and B). Decrease of total IκBα levels and increase of phospho-IκBα suggested that NF-κB signaling was elevated in A20-deficient cells. Consistent with increased NF-κB signaling, expression levels of several NF-κB targets, such as Relb, NF-κB2, TNF, BclX, Hif1α, and Sfpi1, were elevated in the hematopoietic cells of A20Hem-KO mice (Fig. 5 C). These data support the previous findings (Vereecke et al., 2009; Hymowitz and Wertz, 2010; Ma and Malynn, 2012) that NF-κB signaling is augmented in A20-deficient hematopoietic cells.
Figure 5.
Augmented NF-κB activation in A20-deficient hematopoietic cells. (A) Western blot analysis of total and phospho- IκBα in splenocytes either in the presence or absence of LPS stimulation for 1 h. Data are representative of two independent experiments. (B) Quantification of proteins from Western blot shown in A. IκBα protein levels were normalized to Actin protein levels in each lane, respectively. Data are pooled from two independent experiments. (C) Real-time PCR data indicating expression levels of NF-κB target genes in the total BM cells of 14-d-old A20Hem-KO and control mice. Expression levels of target genes were normalized to HPRT levels. Data are representative of three independent experiments. All data represent mean ± SEM. Two-tailed Student’s t tests were used to assess statistical significance (*, P < 0.05).
Increased IFN-γ signaling in A20 mutant HSCs
Earlier studies identified IFN-γ as a direct target of NF-κB proteins (Sica et al., 1997), and based on the critical role of IFN-γ in the maintenance of HSCs (Baldridge et al., 2011; King and Goodell, 2011), we hypothesized that deregulated IFN-γ signaling might be potentially responsible for the HSC phenotype of A20Hem-KO mice. To test this, we measured the expression levels of IFN-γ in A20Hem-KO mice. In line with our hypothesis, we detected elevated levels of Ifn-γ mRNA in both BM and spleen of A20-deficient mice (Fig. 6 A). To substantiate this finding and to identify the cellular source of IFN-γ in A20 mutant mice, we crossed A20Hem-KO mice with the IFN-γ–YFP reporter GREAT (IFN-γ reporter with endogenous polyA transcript) mice (Reinhardt et al., 2009). These transgenic mice have an IRES-eYFP reporter cassette inserted between the translational stop codon and the 3′ UTR/polyA tail of the Ifn-γ gene (Reinhardt et al., 2009). Our analysis of hematopoietic compartments from GREAT-A20Hem-KO mice suggested an elevated expression of YFP and, therefore, IFN-γ in CD11C+, CD3+CD4+, and CD3+CD8+ cells of BM and spleen (Fig. 6 B). In addition, myeloid cells, such as granulocytes and monocytes, from GREAT-A20Hem-KO mice also expressed YFP, albeit at lower levels (Fig. 6 B). Next, we tested whether Ifn-γ is a direct target of NF-κB in A20 mutant BM cells. Our analysis of mouse Ifn-γ gene sequences indicated the presence of five (AAGGACTTCCTC) NF-κB–binding sites (Wong et al., 2011) in its promoter region. Among these sites, one NF-κB–binding site (from −541 to −530) is highly conserved across species (Fig. 6, C and D), and chromatin immunoprecipitation (ChIP) data revealed an increased binding of NF-κB to the Ifn-γ promoter in A20Hem-KO BM cells (Fig. 6 E). To understand the physiological relevance of deregulated IFN-γ expression on the HSC compartment, we investigated whether IFN-γ signaling was augmented in the HSCs from A20Hem-KO mice. Real-time PCR data indicated that the expression levels of IFN-γ target genes, such as Irf1, Irgm1, and STAT1, were elevated in A20 mutant HSCs (Fig. 6 F). Deregulated IFN-γ signaling results in up-regulated surface expression of Sca1 in HSPCs, as Sca1 is a direct target of IFN-γ (Snapper et al., 1991; King and Goodell, 2011). Our analysis of the LSK compartment indicated increased surface expression of Sca1 in hematopoietic progenitors and in total BM cells of A20Hem-KO mice (Fig. 6, G and H; and not depicted). Intriguingly, up-regulation of Sca1 in HSPCs has been connected with increased cell cycle activity (Essers et al., 2009).
Figure 6.
Increased IFN-γ signaling in A20-deficient HSCs. (A) Real-time PCR data indicating expression levels of Ifn-γ transcripts in the BM cells and splenocytes of 14-d-old A20Hem-KO and control mice. Expression levels of Ifn-γ were normalized to Hprt levels. (B) FACS plots indicating YFP expression in the BM cells and splenocytes of IFN-γ–YFP reporter (GREAT) mice, crossed with either control or A20Hem-KO mice. (A and B) Data are representative of three independent experiments. (C) Diagrammatic representation of the Ifn-γ gene indicating the presence of five NF-κB–binding sites in its promoter. (D) NF-κB–binding site in the promoter of Ifn-γ gene of the indicated species. (E) ChIP analysis of NF-κB (p65) binding to the Ifn-γ promoter in the BM cells of 14-d-old A20Hem-KO and control mice. Shown are the real-time PCR data of p65 immunoprecipitates, which were normalized to IgG control immunoprecipitates. Data are representative of two independent experiments. (F) Real-time PCR data indicating expression of IFN-γ target genes in CD150+LSK cells from 14-d-old A20Hem-KO and control mice. Expression levels of target genes were normalized to Hprt levels. Data are pooled from two independent experiments with six mice per group. (G and H) Representative FACS plots (G) and cumulative frequencies (H) indicating expression levels of Sca1 in Lin−c-Kit+CD150+ cells of the BM from 14-d-old A20Hem-KO and control mice. Data are representative of 10 independent experiments (G) and are pooled from six mice per group (H). (I) Real-time PCR data indicating expression levels of IFN-γ–controlled cell cycle regulators in HSCs of 14-d-old A20Hem-KO and control mice. Expression levels of target genes were normalized to Hprt levels. Data are representative of two independent experiments with six mice per group. All data represent mean ± SEM. Two-tailed Student’s t tests were used to assess statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
At the molecular level, a major functional consequence of increased IFN-γ signaling is diminished HSC quiescence, probably caused by deregulated expression of cell cycle regulators, such as CcnD1 and p57, and other key transcriptional targets (de Bruin et al., 2013). Our gene expression experiments identified an increase in the expression levels of CcnD1 (a positive regulator of cell cycle) and a decrease in the expression levels of p57 (a negative regulator of cell cycle) in A20-deficient CD150+LSK cells (Fig. 6 I). Of note, HSCs that are deficient for D-Cyclins show reduced proliferation (Kozar et al., 2004; Pietras et al., 2011), and HSCs lacking p57 failed to reconstitute the hematopoietic system because of loss of quiescence (Matsumoto et al., 2011), a phenotype which is very similar to the phenotype of A20-deficient HSCs. Overall, these data suggest that IFN-γ expression and signaling were elevated in A20Hem-KO mice, and as a consequence, their target genes were up-regulated in HSCs of these animals.
Rescue of the HSC phenotype in A20Hem-KO IFN-γ−/− mice
To directly address the involvement of IFN-γ signaling in the phenotype of A20Hem-KO mice, we crossed A20Hem-KO mice with IFN-γ−/− mice to obtain A20Hem-KO IFN-γ−/− (double KO [DKO]) mice. Strikingly, DKO mice were larger in size and showed more body weight than the A20Hem-KO littermates (Fig. 7, A and B). However, DKO mice were slightly smaller and lighter than the A20F/+ littermate controls (Fig. 7, A and B). Survival curve analysis indicated that postnatal lethality of A20Hem-KO mice was rescued in the DKO mice (Fig. 7 C). Analysis of the HSPC compartment in the BM of DKO mice revealed a rescue of the LSK phenotype (Fig. 7 D). Consistently, the phenotype (relative frequencies) of LSK subsets, including LT-HSCs, ST-HSCs, and MPPs, was rescued in DKO mice (Fig. 7 D). Absolute numbers of LT-HSCs indicated a significant increase (∼300%) of LT-HSC numbers in the DKO mice when compared with their A20Hem-KO littermates (Fig. 7 E). However, when compared with the littermate controls, the numbers of LT-HSCs were still reduced in the DKO mice (Fig. 7 E). In addition, surface expression of Sca1 was restored to WT levels in DKO progenitor cells (Fig. 7 F). Next, we performed BMT experiments to assess the functions of DKO HSCs. Transfer of total BM cells (CD45.2+) from control, IFN-γ−/−, A20Hem-KO, and DKO mice into lethally irradiated WT congenic (CD45.1+) recipients resulted in comparable survival among control, IFN-γ−/−, and DKO recipient groups, whereas mice that received A20Hem-KO BM showed reduced survival (Fig. 7 G). In these particular experiments (Fig. 7 G), we did not observe compromised survival of the recipients that received A20 mutant BM to the extent shown in Fig. 3 A. Nevertheless, analysis of survivors at 5 wk after transplantation indicated undetectable levels of donor cells in recipients that received A20Hem-KO BM cells (Fig. 7 H). In contrast, recipients that received DKO BM cells had ∼40% donor-derived chimera at 5 wk after transplantation (Fig. 7 H), and more importantly, remarkable levels of donor-derived chimera were maintained even at 14 wk after transplantation (Fig. 7 I). To further strengthen these findings, we performed competitive repopulation experiments (donor/competitor ratio = 5:1 and 1:1, respectively) with control, A20Hem-KO, and DKO BM cells. Analysis of recipients indicated detectable levels of donor-derived chimera in both control and DKO groups, whereas donor-derived hematopoiesis was completely absent in recipients that received A20Hem-KO BM (Fig. 7, J and K). Nevertheless, frequencies of donor-derived hematopoiesis were lower in recipients that received DKO BM when compared with the chimera derived from littermate control BM. Donor-derived multilineage analysis indicated intact differentiation of both myeloid and lymphoid lineage cells from DKO BM, whereas it was completely absent in recipients that received A20Hem-KO BM (Fig. 7, L and M). Overall, the data of the DKO mice suggested that the HSC phenotype of A20Hem-KO mice was, at least in part, caused by elevated IFN-γ signaling.
Figure 7.
Rescue of the HSC phenotype in A20Hem-KO IFN-γ−/− mice. (A) Representative images of control, IFN-γ−/−, A20Hem-KO, and A20Hem-KO IFN-γ−/− (DKO) mice showing body size (n = 5). (B) Bar graphs showing body size of control (n = 9), A20Hem-KO (n = 3), and DKO (n = 4) mice. (C) Kaplan-Meier survival curve analysis of control (n = 26), IFN-γ−/− (n = 11), A20Hem-KO (n = 10), and DKO (n = 15) mice. Significance (****, P < 0.0001) was assessed using the log-rank test. (D) FACS plots showing immunophenotype of LSK, LT-HSCs, ST-HSCs, and MPPs of 14-d-old control, IFN-γ−/−, A20Hem-KO, and DKO mice. Data are representative of two independent experiments. (E) Absolute numbers of LT-HSCs in the BM (two femurs and two tibias) of 14-d-old animals. Data are pooled from two independent experiments with two mice per group. (F) Frequencies of Sca1-expressing cells in the Lin−c-Kit+CD150+ fraction of the BM of 14-d-old animals. Data are pooled from two independent experiments with two mice per group. (G) Kaplan-Meier survival curve analysis of WT recipients after BMT. Data are pooled from two independent experiments with 11 mice each for control, A20Hem-KO, and DKO groups and 6 mice for the IFN-γ−/− group. Significance (*, P < 0.03) was assessed using the log-rank test. (H) Frequencies of donor (CD45.2+) cells in the peripheral blood of WT congenic recipients at 5 wk after transplantation. Data are representative of two independent experiments and are pooled from six mice each for control and IFN-γ−/− groups, five mice for the A20Hem-KO group, and seven mice for the DKO group. (I) Frequencies of donor (CD45.2+) cells in the peripheral blood of WT congenic recipients at 14 wk after transplantation. Data are representative of two independent experiments and are pooled from five mice for control and four mice for DKO groups. (J and K) Frequencies of donor (CD45.2+) hematopoiesis in the peripheral blood of WT recipients, at 4 wk after transplantation. Recipients received mixed chimera containing donor (CD45.2+) BM cells (from A20Hem-KO, DKO, and control mice) and WT competitor (CD45.1+) BM cells at a ratio of 1:1 (J) and 5:1 (K), respectively. Data are representative of two independent experiments and are pooled from five mice each for A20Hem-KO and DKO groups and four mice for control group (J) and five mice for DKO group, three mice for A20Hem-KO group, and five mice for control group (K). (L and M) Frequencies of donor-derived cells in total CD11b+ (L) and CD19+ (M) fractions in the peripheral blood of WT recipients from the 1:1 group of J. Data are representative of two independent experiments and are pooled from five mice each for A20Hem-KO and DKO groups and four mice for the control group. All data represent mean ± SEM. Two-tailed Student’s t tests were used to assess statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
In recent years, the ubiquitin-editing enzyme A20 has emerged as a central regulator of inflammation (Liu et al., 2005). Germline deficiency of A20 in mice causes persistent activation of NF-κB, leading to multiorgan inflammation, cachexia, neonatal lethality, and increased responses to TNF and LPS (Lee et al., 2000; Boone et al., 2004). To understand the functions of A20 in the immune system, the Tnfaip3 gene was deleted in selective immune compartments using tissue-specific Cre lines. Disrupting functions of A20 in B cells, using CD19 Cre, resulted in exaggerated NF-κB signaling and hyperresponsiveness to LPS/B cell receptor and CD40 ligation. In addition, A20 deficiency in B cells caused elevated levels of IL6 expression, spontaneous activation of B cells, expansion of myeloid cells, plasma cell hyperplasia, and autoimmunity at 3–4 mo of age (Tavares et al., 2010; Chu et al., 2011). DC-specific deletion of A20, using CD11c Cre mice, resulted in spontaneous activation of DCs (which results in increased secretion of proinflammatory cytokines), B cells, T cells, and myeloid cells and development of lymphadenopathy and splenomegaly in 4–6-wk-old mice (Hammer et al., 2011; Kool et al., 2011). Macrophage- and granulocyte-specific deletion of A20, using LysM Cre mice, caused spontaneous polyarthritis, increased systemic and local cytokine production, and increased osteoclast functions (Matmati et al., 2011). Although these studies have elegantly demonstrated the importance of A20-mediated NF-κB down-regulation in the immune system, almost all of these studies were focused on the role of A20 on the functions of immune cells. In the present study, we focused on the role of A20 in early hematopoiesis and found that A20 deficiency results in hyperproliferation of HSPCs and in premature depletion of the HSC pool. Thus, A20 acts as a key modulator in the maintenance of HSC quiescence, particularly under steady-state conditions.
The role of infection and inflammation in the control of hematopoiesis has gained a lot of attention in the recent years. Although it was believed that only lineage-committed progenitors, including CMP and CLP, were involved in sensing and in responding to infections (Welner et al., 2008; Baldridge et al., 2011), it is now evident that HSCs are directly involved in the primary response of both acute and chronic infections (King and Goodell, 2011). The direct response of HSCs to an immune insult might be a key determinant in the clearance of the pathogen because HSCs can dictate cell fate decisions and can preferentially differentiate into a particular hematopoietic lineage to clear the pathogen. HSCs have evolved to sense pathogens by both cell-intrinsic and -extrinsic mechanisms (Baldridge et al., 2011; King and Goodell, 2011). HSPCs have been shown to recognize pathogens in a cell-intrinsic manner, through their expression of TLRs. In contrast, the cell-extrinsic mode of recognition of an immune insult by HSCs primarily involves expression and stimulation by proinflammatory cytokines and chemokines, which includes IL6, TNF, CC-chemokine ligand 2 (CCL2), IFN-α, and IFN-γ (Baldridge et al., 2011; King and Goodell, 2011).
Indeed, long-term treatment of mice with low doses of LPS resulted in impaired self-renewal and multilineage differentiation of HSCs (Esplin et al., 2011). Exposure of HSCs to a combination of IL6, TNF, and CCL2, a condition which mimics LPS stimulation, resulted in defective long-term engraftment (Chen et al., 2010).
Recent studies identified IFNs, both type I (IFN-α) and type II (IFN-γ), as major regulators of HSC quiescence. IFNs are produced by lymphocytes, such as NK cells, NKT cells, and T cells, during bacterial and viral infections. Although in vitro stimulation of HSCs with IFN-α has antiproliferative effects, in vivo treatment of mice with IFN-α results in loss of HSC quiescence, increased phosphorylation of STAT1 and AKT1, expression of IFN-α target genes, up-regulation of surface Sca1, and increased cell cycle activity mediated by Sca1 (Essers et al., 2009). Interestingly, transient activation of HSCs by IFN-α did not affect the numbers of functional HSCs. However, long-term (chronic) activation of the IFN-α pathway in HSCs leads to decreased HSC activity and a marked competitive disadvantage in mixed BM reconstitution experiments (Essers et al., 2009). Consistently, loss of IFN regulatory factor-2 (IRF-2), a transcriptional suppressor of IFN signaling, results in reduced HSC pool, diminished HSC quiescence, and compromised HSC functions as the result of increased IFN-α signals (Sato et al., 2009). In contrast, stimulation of HSCs with IFN-γ leads to hyperproliferation of HSCs both in vitro and in vivo (Baldridge et al., 2010), and IFN-γ has been shown to be sufficient to promote proliferation of long-term repopulating HSCs, accompanied by a remarkable increase of immediate downstream progenitors (Baldridge et al., 2010). This study found a disadvantage in competitive repopulation assays after Mycobacterium avium infection (IFN-γ exposure) and proposed that baseline IFN-γ tone may influence HSC turnover and quality during normal homeostasis (Baldridge et al., 2010). Consistently, King et al. (2011) showed that dysregulated IFN-γ signaling results in reduction of the HSC pool. Overall, our data agree with the existing paradigm of IFN signaling in HSCs (Essers et al., 2009; King et al., 2011; Pietras et al., 2014). The current study indicated that IFN-γ signaling has an indispensable role in the hematopoietic phenotype of A20-deficient mice. However, analysis of A20–IFN-γ DKO mice indicated only a partial rescue of the A20Hem-KO phenotype. These data suggest that IFN-γ is one of the several factors contributing to the HSC phenotype of A20Hem-KO mice. Other potential factors that could influence the phenotype of A20-deficient mice may include cytokines, such as type I IFNs, IL6, and TNF. Indeed, earlier studies identified critical roles of these cytokines in the control of HSCs and hematopoiesis (Baldridge et al., 2011; King and Goodell, 2011; Pietras et al., 2011). Among these cytokines, it would be more interesting to study the role of type I IFNs, as mounting evidence documents the direct involvement of A20 in the negative regulation of two key inducers of type I IFN, IRF3 and IRF7 (Hymowitz and Wertz, 2010; Parvatiyar and Harhaj, 2011).
Further studies that aim to dissect the potential involvement of other proinflammatory cytokines in A20-deficient mice would be helpful in understanding the role of inflammation in HSC biology. We anticipate that our study contributes to a deeper understanding of hematological disorders in patients with TNFAIP3 (A20) mutations.
MATERIALS AND METHODS
Mice.
To generate Tnfaip3 conditional KO mice, the mouse Tnfaip3 exon 3 region and homology arms were PCR amplified from C57BL/6 mouse genomic DNA and cloned into the SalI, XhoI, and SfiI sites of the pEasy-FRTlox DT vector (provided by K. Rajewsky, Harvard Medical School, Boston, MA), which contains a neomycin resistance cassette for positive selection and a diphtheria toxin cassette for negative selection. The targeting vector was linearized by ClaI and electroporated into Bruce4 embryonic stem cells. G418-resistant clones were screened for homologous recombination by Southern blotting. Correctly targeted embryonic stem clones were injected into albino C57BL/6-cBrd/cBrd blastocysts (National Cancer Institute, Frederick, MD) and implanted into host female mice to generate chimeras. Chimeras were then bred to albino C57BL/6 mice to obtain black C57BL/6 litters, which were then screened for germline transmission of the allele. Mice were crossed to the ACTB-Flpe mouse strain (The Jackson Laboratory) to remove the neomycin cassette. VAVCre/+, IFN-γ−/−, and IFN-γ reporter (GREAT) mice were purchased from The Jackson Laboratory. CD45.1 congenic animals were purchased from the National Cancer Institute. In our experiments, we used A20+/+, A20F/+, A20F/F, and A20F/+ VavCre/+ mice as controls, as none of these control animals exhibited any hematopoietic phenotype. However, in each experiment, age-and gender-matched littermates were used as controls. Mice were maintained under specific pathogen–free conditions and were used according to the protocols approved by the Institutional Animal Care and Use Committee of Columbia University Medical Center.
Cell preparation.
A20Hem-KO mice were analyzed at 14 d after birth (P14). RBCs were lysed with ammonium chloride (STEMCELL Technologies). Trypan blue (Amresco)–negative cells were counted as live cells.
Flow cytometry.
Cells were analyzed by flow cytometry with either LSRFortessa or LSRII (both BD) and FACSDiva software (BD) or FlowJo software (Tree Star). Cell sorting was performed with a FACSAria (BD). The following monoclonal antibodies were used: anti-B220 (RA3-6B2), anti-CD19 (1D3), anti-CD3ε (145-2C11), anti-CD4 (GK1.5), anti-CD8 (53-6.7), anti-CD11b (M1/70), anti-CD34 (RAM34), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD48 (HM48-1), anti-CD117 (2B8), anti-Flt3 (A2F10.1), anti–Gr-1 (RB6-8C5), anti–Sca-1 (D7), and anti-Ter119 (TER119; from BD); anti-CD150 (TC15-12F12.2; from BioLegend); and anti-CD16/32 (93) and anti-CD127 (A7R34; from eBioscience). Cells incubated with biotinylated monoclonal antibodies were incubated with fluorochrome-conjugated streptavidin–peridinin chlorophyll protein–cyanine 5.5 (551419; BD) or streptavidin-allophycocyanin-Cy7 (554063; BD). In all of the FACS plots, the relative frequencies (%) of the gated fraction are indicated.
BMT experiments.
For radioprotection assays, 106 of BM cells were injected into lethally irradiated (10 Gy) congenic (CD45.1+) recipient mice. For competitive repopulation experiments, 4 × 105, 1.2 × 106, or 106 of BM cells from either A20 control or A20Hem-KO mice were mixed with 4 × 105, 4 × 105, or 2 × 105 of WT (CD45.1+) BM cells (to obtain a ratio of 1:1, 3:1, or 5:1, respectively) and were injected into lethally irradiated congenic WT (CD45.1+) recipient mice. For serial transplantation, 6 × 105 BM cells from the primary transplants were injected into lethally irradiated congenic WT recipients.
Apoptosis assays.
Apoptotic cells were detected with the Annexin V PE Apoptosis Detection kit or Annexin V FITC Apoptosis Detection kit (BD) according to the manufacturer’s instructions. For TUNNEL assay, the In Situ Cell Death Detection kit (Roche) was used according to the manufacturer’s instructions.
Cell proliferation experiments.
For BrdU assays, either 14-d-old (P14) mice or 1-d-old (P1) pups were injected i.p. with either 1 Mg or 200 µg BrdU (BD), respectively. At 16 h after injection, mice/pups were sacrificed and BM/liver cells were stained for BrdU, according to the BrdU Flow kit (BD) manufacturer’s instructions, and detected by flow cytometry. For Ki-67 analysis, BM cells were stained for surface markers and fixed and permeabilized with FIX & PERM Cell Fixation & Cell Permeabilization kit (Life Technologies). These cells were further stained with anti–Ki-67 antibodies (B56; BD).
SP assays.
BM cells were incubated with 5 µg/ml Hoechst 33342 (Life Technologies) at 37°C for 90 min.
Cell cycle analysis.
For Pyronin Y analysis, cells were first incubated with 5 µg/ml Hoechst 33342 at 37°C for 45 min and then with 1 µg/ml Pyronin Y (Sigma-Aldrich) for an additional 45 min at 37°C. For cell cycle assays using Ki-67 and Hoechst, cells were stained with surface markers and fixed and permeabilized with Cytofix/Cytoperm fixation/permeabilization solution kit (BD). These cells were stained with Ki-67 for 30 min and subsequently stained with 20 µg/ml Hoechst 33342 for 5 min and analyzed by flow cytometry.
Homing assay.
LSK cells were sorted from the BM and stained with CFSE (Life Technologies) for 15 min, and 5 × 104 CFSE+LSK cells were injected i.v. into lethally irradiated (10 Gy) WT mice. 20 h after injection, mice were sacrificed, and CFSE-positive cells in the BM were analyzed by flow cytometry.
Western blot analysis.
For immunoblot analyses, cells were lysed with cell lysis buffer (Cell Signaling Technology) with protease inhibitor cocktail (Complete; Roche) and 1 mM PMSF (Santa Cruz Biotechnology, Inc.). Cell lysates were boiled with sample buffer (NuPAGE; Life Technologies) containing 1% 2-Mercaptoethanol (Sigma-Aldrich). Proteins were subjected to 8–12% SDS-PAGE and transferred to PVDF membranes (Bio-Rad Laboratories). The membranes were blocked with either 5% bovine serum albumin (Life Technologies) or 5% skim milk and then treated with primary and secondary antibodies, respectively. The blots were visualized using the ProtoGlow ECL (National Diagnostics) and Image station 440 (Kodak). Antibodies used were as follows: anti-A20 (D13H3; Cell Signaling Technology), anti-IκBα (44D4; Cell Signaling Technology), anti–phospho-IκBα (5A5; Cell Signaling Technology), anti-actin (I-19; Santa Cruz Biotechnology, Inc.), HRP-conjugated anti–mouse and anti–rabbit IgG (Cell Signaling Technology), and HRP-conjugated anti–goat IgG (Santa Cruz Biotechnology, Inc.).
IκBα degradation assay.
Splenocytes were stimulated with 1 µg/ml LPS (Sigma-Aldrich) at 37°C for 1 h, and then lysed for protein extraction. Cells with no stimulation served as controls.
ChIP assay.
ChIP assay was performed with an Agarose ChIP kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. In brief, 107 of BM cells were fixed and immunoprecipitated with either anti-p65 (D14E12; Cell Signaling Technology) or rabbit IgG (Thermo Fisher Scientific) antibodies. Immunoprecipitated DNA fragments were quantified by real-time PCR with the use of the following primers, which amplify the IFN-γ promoter region containing NF-κB–binding sites: forward, 5′-ACCCTGAGTGATTTGTAGTAGGT-3′; and reverse, 5′-GTGAATTTCTCATCCACAGAGCA-3′. Fold enrichment was normalized to rabbit IgG–precipitated samples.
RNA extraction, PCR, and real-time PCR.
Total RNA was isolated with an RNeasy Mini kit (QIAGEN). cDNA was synthesized with Oligo(dT) primer and Maxima reverse transcription (Thermo Fisher Scientific). PCR was performed with T100 thermal cycler (Bio-Rad Laboratories) and TSG Taq (Lamda Biotech). Real-time PCR was performed in duplicates with a CFX-connect real-time PCR system (Bio-Rad Laboratories) and SsoAdvanced SYBR Green Supermix (Bio-Rad Laboratories) according to the manufacturer’s instructions. Relative expression was normalized to the expression levels of the internal control (housekeeping gene) Hprt.
Statistical analyses.
We used the unpaired Student’s t test for all the experiments, except survival experiments. For survival curve analysis, log-rank test was used. Differences with P < 0.05 were considered statistically significant and are denoted as *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Acknowledgments
We thank Riccardo Dalla-Favera for expert advice and critical reading of the manuscript and Danielle Matsushima for help with correcting the manuscript.
The authors declare no competing financial interests.
Author contributions: M.M. Nakagawa performed all of the experiments, analyzed and interpreted data, and prepared the figures. K. Thummar helped with establishing the animal colony. J. Mandelbaum and L. Pasqualucci generated the A20Floxed mice. C.V. Rathinam conceived, designed, and directed research, interpreted data, provided advice, and wrote the manuscript. All authors discussed and reviewed the manuscript.
Footnotes
Abbreviations used:
- BMT
- BM transplantation
- ChIP
- chromatin immunoprecipitation
- DKO
- double KO
- DUB
- deubiquitinase
- GREAT
- IFN-γ reporter with endogenous polyA transcript
- HSC
- hematopoietic stem cell
- HSPC
- hematopoietic stem and progenitor cell
- LSK
- Lineage−Sca1+c-Kit+
- LT-HSC
- long-term HSC
- MEF
- mesenchymal embryonic fibroblast
- MPP
- multipotent progenitor
- PTM
- posttranslational modification
- SP
- side population
- ST-HSC
- short-term HSC
References
- Baldridge M.T., King K.Y., Boles N.C., Weksberg D.C., and Goodell M.A.. 2010. Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature. 465:793–797 10.1038/nature09135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge M.T., King K.Y., and Goodell M.A.. 2011. Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 32:57–65 10.1016/j.it.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone D.L., Turer E.E., Lee E.G., Ahmad R.C., Wheeler M.T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., et al. . 2004. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 5:1052–1060 10.1038/ni1110 [DOI] [PubMed] [Google Scholar]
- Chen C., Liu Y., Liu Y., and Zheng P.. 2010. Mammalian target of rapamycin activation underlies HSC defects in autoimmune disease and inflammation in mice. J. Clin. Invest. 120:4091–4101 10.1172/JCI43873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Rodrigues N., Shen H., Yang Y., Dombkowski D., Sykes M., and Scadden D.T.. 2000. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 287:1804–1808 10.1126/science.287.5459.1804 [DOI] [PubMed] [Google Scholar]
- Chu Y., Vahl J.C., Kumar D., Heger K., Bertossi A., Wójtowicz E., Soberon V., Schenten D., Mack B., Reutelshöfer M., et al. . 2011. B cells lacking the tumor suppressor TNFAIP3/A20 display impaired differentiation and hyperactivation and cause inflammation and autoimmunity in aged mice. Blood. 117:2227–2236 10.1182/blood-2010-09-306019 [DOI] [PubMed] [Google Scholar]
- de Bruin A.M., Demirel Ö., Hooibrink B., Brandts C.H., and Nolte M.A.. 2013. Interferon-γ impairs proliferation of hematopoietic stem cells in mice. Blood. 121:3578–3585 10.1182/blood-2012-05-432906 [DOI] [PubMed] [Google Scholar]
- Esplin B.L., Shimazu T., Welner R.S., Garrett K.P., Nie L., Zhang Q., Humphrey M.B., Yang Q., Borghesi L.A., and Kincade P.W.. 2011. Chronic exposure to a TLR ligand injures hematopoietic stem cells. J. Immunol. 186:5367–5375 10.4049/jimmunol.1003438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., and Trumpp A.. 2009. IFNα activates dormant haematopoietic stem cells in vivo. Nature. 458:904–908 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Goodell M.A., Brose K., Paradis G., Conner A.S., and Mulligan R.C.. 1996. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 183:1797–1806 10.1084/jem.183.4.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer G.E., Turer E.E., Taylor K.E., Fang C.J., Advincula R., Oshima S., Barrera J., Huang E.J., Hou B., Malynn B.A., et al. . 2011. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat. Immunol. 12:1184–1193 10.1038/ni.2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitotsumatsu O., Ahmad R.C., Tavares R., Wang M., Philpott D., Turer E.E., Lee B.L., Shiffin N., Advincula R., Malynn B.A., et al. . 2008. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 28:381–390 10.1016/j.immuni.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H., Hamblen M.J., Rooke H.M., Schindler J.W., Saleque S., Fujiwara Y., and Orkin S.H.. 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 431:1002–1007 10.1038/nature02994 [DOI] [PubMed] [Google Scholar]
- Hymowitz S.G., and Wertz I.E.. 2010. A20: from ubiquitin editing to tumour suppression. Nat. Rev. Cancer. 10:332–341 10.1038/nrc2775 [DOI] [PubMed] [Google Scholar]
- King K.Y., and Goodell M.A.. 2011. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11:685–692 10.1038/nri3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K.Y., Baldridge M.T., Weksberg D.C., Chambers S.M., Lukov G.L., Wu S., Boles N.C., Jung S.Y., Qin J., Liu D., et al. . 2011. Irgm1 protects hematopoietic stem cells by negative regulation of IFN signaling. Blood. 118:1525–1533 10.1182/blood-2011-01-328682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M., van Loo G., Waelput W., De Prijck S., Muskens F., Sze M., van Praet J., Branco-Madeira F., Janssens S., Reizis B., et al. . 2011. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity. 35:82–96 10.1016/j.immuni.2011.05.013 [DOI] [PubMed] [Google Scholar]
- Kozar K., Ciemerych M.A., Rebel V.I., Shigematsu H., Zagozdzon A., Sicinska E., Geng Y., Yu Q., Bhattacharya S., Bronson R.T., et al. . 2004. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 118:477–491 10.1016/j.cell.2004.07.025 [DOI] [PubMed] [Google Scholar]
- Lee E.G., Boone D.L., Chai S., Libby S.L., Chien M., Lodolce J.P., and Ma A.. 2000. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 289:2350–2354 10.1126/science.289.5488.2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.C., Penninger J., and Karin M.. 2005. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 5:941–952 10.1038/nri1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A., and Malynn B.A.. 2012. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat. Rev. Immunol. 12:774–785 10.1038/nri3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matmati M., Jacques P., Maelfait J., Verheugen E., Kool M., Sze M., Geboes L., Louagie E., Mc Guire C., Vereecke L., et al. . 2011. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat. Genet. 43:908–912 10.1038/ng.874 [DOI] [PubMed] [Google Scholar]
- Matsumoto A., Takeishi S., Kanie T., Susaki E., Onoyama I., Tateishi Y., Nakayama K., and Nakayama K.I.. 2011. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 9:262–271 10.1016/j.stem.2011.06.014 [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K., Reavie L.B., and Aifantis I.. 2012. Regulation of hematopoietic stem cell fate by the ubiquitin proteasome system. Trends Immunol. 33:357–363 10.1016/j.it.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., Uchida N., and Weissman I.L.. 1995. The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol. 11:35–71 10.1146/annurev.cb.11.110195.000343 [DOI] [PubMed] [Google Scholar]
- Ogilvy S., Metcalf D., Gibson L., Bath M.L., Harris A.W., and Adams J.M.. 1999. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 94:1855–1863. [PubMed] [Google Scholar]
- Orford K.W., and Scadden D.T.. 2008. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9:115–128 10.1038/nrg2269 [DOI] [PubMed] [Google Scholar]
- Orkin S.H., and Zon L.I.. 2008. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 132:631–644 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar K., and Harhaj E.W.. 2011. Regulation of inflammatory and antiviral signaling by A20. Microbes Infect. 13:209–215 10.1016/j.micinf.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué E., Wagers A.J., Giuriato S., Anderson W.C., and Weissman I.L.. 2005. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 202:1599–1611 10.1084/jem.20050967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras E.M., Warr M.R., and Passegué E.. 2011. Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 195:709–720 10.1083/jcb.201102131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras E.M., Lakshminarasimhan R., Techner J.M., Fong S., Flach J., Binnewies M., and Passegué E.. 2014. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 211:245–262 10.1084/jem.20131043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam C., and Flavell R.A.. 2010. c-Cbl deficiency leads to diminished lymphocyte development and functions in an age-dependent manner. Proc. Natl. Acad. Sci. USA. 107:8316–8321 10.1073/pnas.0914496107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam C., Thien C.B., Langdon W.Y., Gu H., and Flavell R.A.. 2008. The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev. 22:992–997 10.1101/gad.1651408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam C., Thien C.B., Flavell R.A., and Langdon W.Y.. 2010. Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell. 18:341–352 10.1016/j.ccr.2010.09.008 [DOI] [PubMed] [Google Scholar]
- Rathinam C., Matesic L.E., and Flavell R.A.. 2011. The E3 ligase Itch is a negative regulator of the homeostasis and function of hematopoietic stem cells. Nat. Immunol. 12:399–407 10.1038/ni.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt R.L., Liang H.E., and Locksley R.M.. 2009. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10:385–393 10.1038/ni.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Onai N., Yoshihara H., Arai F., Suda T., and Ohteki T.. 2009. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat. Med. 15:696–700 10.1038/nm.1973 [DOI] [PubMed] [Google Scholar]
- Sica A., Dorman L., Viggiano V., Cippitelli M., Ghosh P., Rice N., and Young H.A.. 1997. Interaction of NF-κB and NFAT with the interferon-γ promoter. J. Biol. Chem. 272:30412–30420 10.1074/jbc.272.48.30412 [DOI] [PubMed] [Google Scholar]
- Snapper C.M., Yamaguchi H., Urban J.F. Jr, and Finkelman F.D.. 1991. Induction of Ly-6A/E expression by murine lymphocytes after in vivo immunization is strictly dependent upon the action of IFN-α/β and/or IFN-γ. Int. Immunol. 3:845–852 10.1093/intimm/3.9.845 [DOI] [PubMed] [Google Scholar]
- Sun S.C.2008. Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 8:501–511 10.1038/nri2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares R.M., Turer E.E., Liu C.L., Advincula R., Scapini P., Rhee L., Barrera J., Lowell C.A., Utz P.J., Malynn B.A., and Ma A.. 2010. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 33:181–191 10.1016/j.immuni.2010.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A., Essers M., and Wilson A.. 2010. Awakening dormant haematopoietic stem cells. Nat. Rev. Immunol. 10:201–209 10.1038/nri2726 [DOI] [PubMed] [Google Scholar]
- Vereecke L., Beyaert R., and van Loo G.. 2009. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 30:383–391 10.1016/j.it.2009.05.007 [DOI] [PubMed] [Google Scholar]
- Welner R.S., Pelayo R., Nagai Y., Garrett K.P., Wuest T.R., Carr D.J., Borghesi L.A., Farrar M.A., and Kincade P.W.. 2008. Lymphoid precursors are directed to produce dendritic cells as a result of TLR9 ligation during herpes infection. Blood. 112:3753–3761 10.1182/blood-2008-04-151506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I.E., O’Rourke K.M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D.L., et al. . 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 430:694–699 10.1038/nature02794 [DOI] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E., et al. . 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 135:1118–1129 10.1016/j.cell.2008.10.048 [DOI] [PubMed] [Google Scholar]
- Wong D., Teixeira A., Oikonomopoulos S., Humburg P., Lone I.N., Saliba D., Siggers T., Bulyk M., Angelov D., Dimitrov S., et al. . 2011. Extensive characterization of NF-κB binding uncovers non-canonical motifs and advances the interpretation of genetic functional traits. Genome Biol. 12:R70 10.1186/gb-2011-12-7-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]