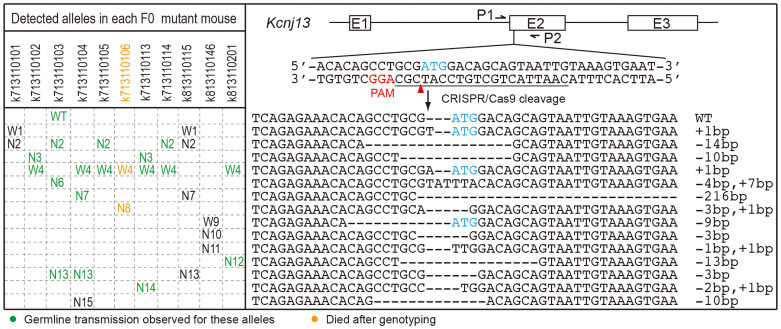
Figure 1. CRISPR-Cas9-induced mutations in Kcnj13 F0 mice.
E1, E2 and E3 in the schematic of the Kcnj13 gene structure indicate three known exons. P1 and P2 are genotyping primers for the CRISPR-Cas9 targeting site. The blue ATG is the putative start codon of Kcnj13. The red arrowhead indicates the potential Cas9 cleavage site. PAM: the protospacer-adjacent motif required for the binding and cleavage of DNA by CRISPR-Cas9. The underlined sequence is the sgRNA target we selected to delete the start codon. Small deletions (1–14 bp), small insertions (1 and 7 bp), and one large deletion (216 bp) were detected. All ATG-deleted alleles are defined as candidate Kcnj13 null alleles (N, named N2-15); alleles that leave the ATG intact are defined as wild-type alleles (W, named W1, W4 and W9). The wild-type reference allele is abbreviated as “WT”. Seven F0 mutant mice were selected to intercross or cross with wild-type C57BL/6 mice to test if their mutant alleles could be transmitted to F1 animals; transmitted alleles are labeled in green.