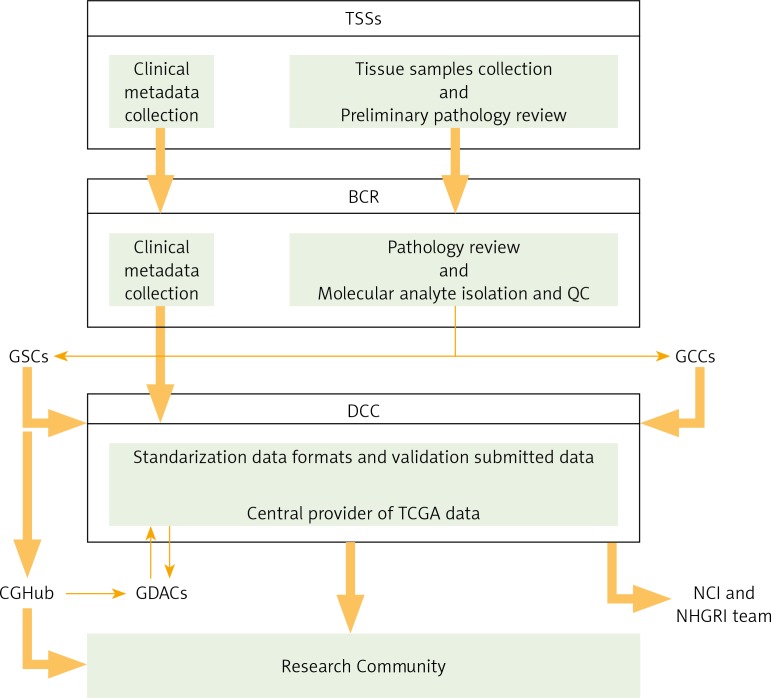
Fig. 1.
The Cancer Genome Atlas (TCGA) Research Network Centres flowchart. Based on [6]
The TCGA structure involves several cooperating centres for processing the samples and managing all the obtained datasets. First, different Tissue Source Sites (TSSs) collect clinical metadata and biospecimens from eligible cancer patients. After preliminary pathology review, TSSs deliver biospecimens and metadata to the Biospecimen Core Resource (BCR), where they are approved. Next, the BCR catalogues and submits metadata to the Data Coordinating Centre (DCC), as well as processing and verifying the quality and quantity of isolated molecular analytes, which are further provided to Genome Characterisation Centres (GCCs) and Genome Sequencing Centres (GSCs) for further genomic characterisation and high-throughput sequencing. Then, sequence-related data are deposited in DCC. The GSCs also submit trace files, sequences, and alignment mappings to NCI's Cancer Genomics Hub (CGHub) secure repository. Generated genomic data submitted to the DCC and CGHub are made available to the research community and Genome Data Analysis Centres (GDACs). The GDACs provide new information-processing, analysis, and visualisation tools to the entire research community. Furthermore, the information generated by the TCGA Research Network is centrally managed at the DCC and entered into public free-access databases (TCGA Portal, NCBI's Trace Archive, CGHub), allowing scientists to continually access the cancer datasets