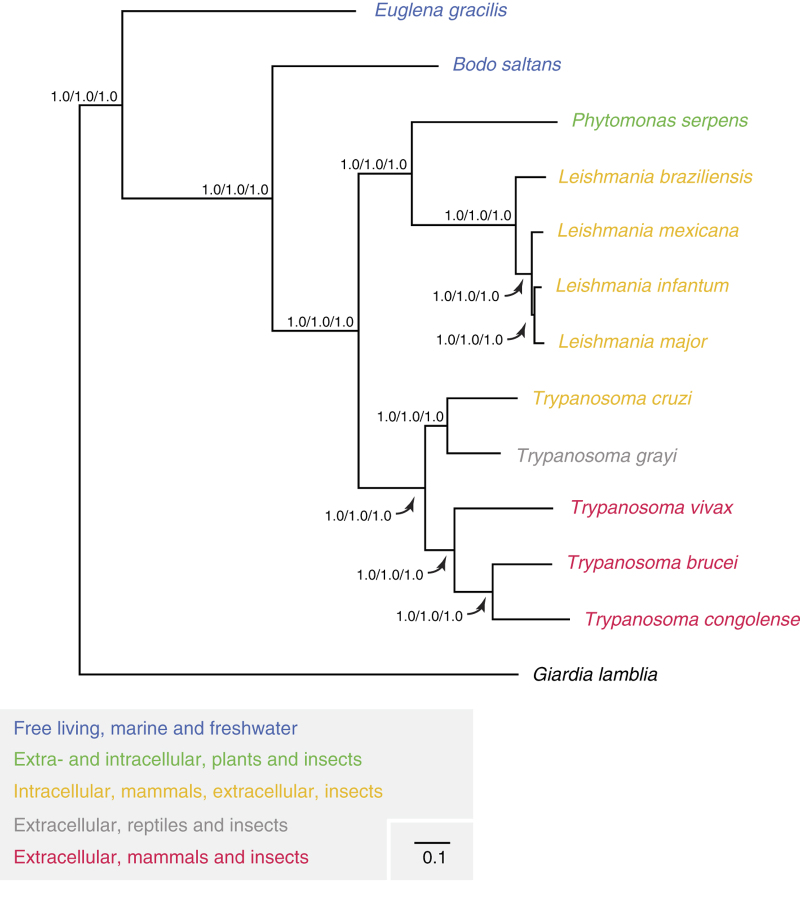
Figure 1.
Phylogenetic tree of selected Euglenozoa. Phylogeny is inferred from 119,006 aligned amino acid positions (1,547,078 amino acids) from 959 nuclear genes. The topology was calculated using Bayesian inference (BI), bootstrapped maximum likelihood (ML) analysis and bootstrapped neighbor joining (NJ). ML used the JTT model of amino acid substitution and CAT rates, BI used the WAG model of amino acid substitution and gamma distributed rates approximated by four discrete gamma categories. Branch lengths shown are from the ML topology, scale bar indicates number of changes per site. Values shown at internal nodes represent bootstrap support values for ML and NJ tree as well as posterior probabilities for BI tree. In all cases support for each bipartition in the topology is 100%. Giardia lamblia, a diplomonad excavate, is included as an outgroup as the Euglenozoa are also members of the Excavata.