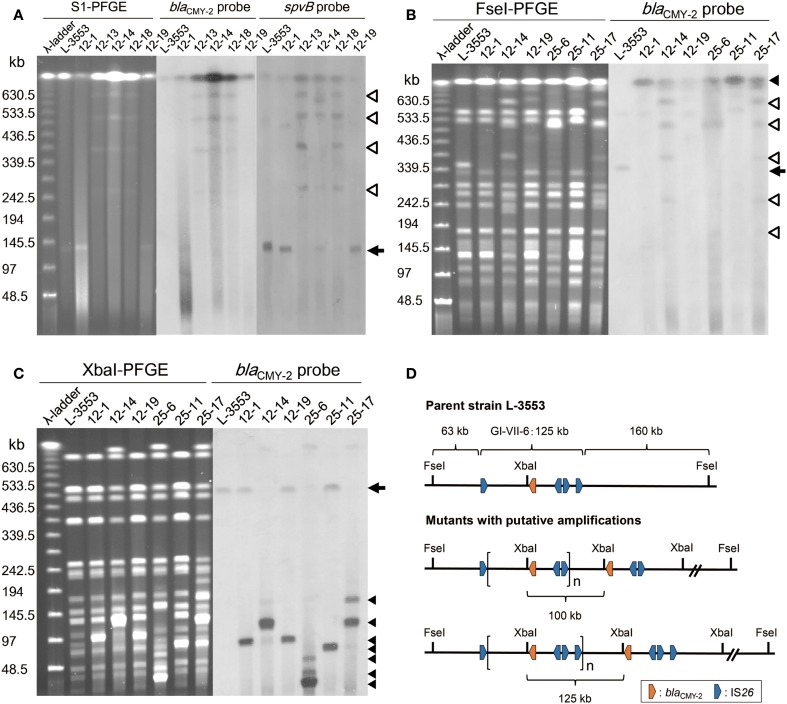
Figure 4.
PFGE–Southern hybridization images demonstrating blaCMY−2 and spvB locations in S. Typhimurium strains. (A) PFGE separation of S1 nuclease-digested genomic DNA from the selected strains followed by Southern hybridization with probes specific to blaCMY−2 and spvB. An arrow indicates the position of the endogenous 133-kb plasmid, in which the spvB signal was detected. Open triangles indicate the positions of enlarged plasmids, in which both blaCMY−2 and spvB signals were detected. Both signals were observed in multiple plasmid bands in strains 12-13, 12-14, and 12-18. (B) PFGE separation of FseI-digested genomic DNA from the selected strains followed by Southern hybridization with a blaCMY−2 probe. An arrow indicates the original position of the restricted fragment (350 kb) in which the chromosomal blaCMY−2 signal was detected. In the selected mutants, this fragment was not present and the blaCMY−2 signal was identified on the largest band as indicated by a closed trangle. Open triangles indicate the position of enlarged plasmid observed in strains 12-14, 25-6, and 25-17. (C) PFGE separation of XbaI-digested genomic DNA from the selected strains followed by Southern hybridization with a blaCMY−2 probe. An arrow indicates the original position of the restricted fragment (520 kb) in which chromosomal blaCMY−2 signal was detected. Closed triangles indicate the position of blaCMY−2 signals that originated from tandemly amplified GI-VII-6. (D) Restriction maps of the GI-VII-6 and its flanking region in the parent strain L-3553 and the mutants with putative amplifications. One putative amplified region presented is flanked by the first and the third copies of IS26 (upper), and the other is flanked by the first and the fourth copies of IS26 (lower). These amplified regions generate 100-kb (upper) and 125-kb (lower) fragments after XbaI digestion, respectively.