Abstract
The DNA-binding specificity of a transcription factor, the FFRP FL4 (pot1613368) from Pyrococcus sp. OT3, was studied. Using SELEX (systematic evolution of ligands by exponential environment) experiments, from a set of fragments, ∼150 bps, of the genomic DNA of P. OT3, seven were selected as containing binding sites. Thirteen bases identified as shared by the seven selected fragments with the least mismatches, 2.71 on average, was ATGAAAAAGTCAT. This sequence was closely related with another sequence, ATGAA[AAA/TTT]TTCAT, in the 5-3-5 arrangement, i.e. NANBNCNDNE[AAA/TTT]NENDNCNBNA, where, e.g. NA was the base complementary to NA. The average number of mismatches found between this sequence and the seven fragments was 3.14. A sequence, TTGAAATTTACAA, resembling the sequence ATGAA[AAA/TTT]TTCAT and also another 5-3-5 sequence, TTGAA[AAA/TTT]TTCAA, was found upstream of the fl4 gene, which is potentially recognized by FL4 for auto-regulation. Thus it is likely that an ideal binding-site of FL4 is ATGAA[AAA/TTT]TTCAT or TTGAA[AAA/TTT]TTCAA. In this abstract, the sequences were highlighted in Italic at 3, and with bold characters at 5 and 5. When two sequences compared were the same at some positions, there they were underlined.
Keywords: Archaea, AsnC, DNA-protein interaction, hyper-thermophile, Lrp, transcription factor
Introduction
We have been studying the structure and function of feast/famine regulatory proteins (FFRPs), regulating transcription of many genes in archaea and eubacteria.1)–25) The nucleotide sequences of DNA sites bound by various FFRPs are summarized into the same form, NANBNCNDNE[AAA/TTT]NENDNCNBNA, where, e.g. NA is the base complementary to NA1),4),18),22): here referred to as the 5-3-5 arrangement. In the reverse direction, several transcription factors experimentally identified as binding DNA sequences in this arrangement have been re-identified as FFRPs by analyzing their amino acid sequences carefully.4) Thus the 5-3-5 arrangement appears to be uniquely associated with FFRPs.
When 13 bps in the 5-3-5 arrangement are positioned by overlapping onto a TATA box or its downstream, transcription of the gene will be repressed by binding by an FFRP.1),9) While, when such 13 bps are positioned immediately upstream of a TATA box with an insertion of ∼4 or ∼15 bps (i.e. 4 plus 10.5), binding of an FFRP will activate transcription, possibly through its interaction with the TATA-binding protein (TBP), thereby recruiting TBP to the TATA box.1),5),9)
In this paper, results obtained by SELEX (systematic evolution of ligands by exponential environment)26) experiments are analyzed in order to determine the DNA-binding specificity of an FFRP, FL4 (pot1613368) from a hyper-thermophilic archaeon, Pyrococcus sp. OT3. This FFRP is one of sixteen FFRPs we have identified as coded in the genome27) of this organism (Table I). The experiments which we report in this paper were, in fact, carried out several years ago. Yet many possible consensus sequences can be deduced from seven fragments of ∼270 bps each selected, and so by statistical analyses alone we were unable to pinpoint a short consensus sequence uniquely. Only recently by assuming a 5-3-5 arrangement for FL4-binding sites, we have come to a conclusion.
Table I.
Transcription factors, FFRPs, identified as coded in the genome of Pyrococcus sp. OT3
| ID | Fuller ID* | Orthologues from other species | Crystal 3D | EM analysis | Ligand | Binding DNA-sequences |
|---|---|---|---|---|---|---|
| DM1 | pot1216151 | none | + | + | isoleucine | N.A. |
| DM2 | pot0300646 | none | N.D. | N.E. | N.D. | N.A. |
| DM3 | pot0175330 | none | N.D. | N.E. | N.D. | N.A. |
| FL1 | pot0828564 | none | N.D. | N.E. | N.D. | N.D. |
| FL2 | pot0836696 | none | N.D. | N.E. | N.D. | N.D. |
| FL3 | pot0868477 | none | N.D. | N.E. | N.D. | N.D. |
| FL4 | pot1613368 | none | N.D. | N.E. | N.D. | ATGAA** |
| FL5 | pot1664679 | none | N.D. | N.E. | N.D. | N.D. |
| FL6 | pot1735659 | none | N.D. | N.E. | N.D. | N.D. |
| FL7 | pot0008824 | none | N.D. | N.E. | N.D. | N.D. |
| FL8 | pot0123002 | none | N.D. | N.E. | N.D. | N.D. |
| FL9 | pot0301583 | none | N.D. | N.E. | N.D. | N.D. |
| FL10 | pot0377090 | LrpA from P. f. | + | − | N.D. | TTCG2) |
| FL11 | pot0434017 | none | + | + | (glutamine) | TGAAA6) |
| FL12 | pot0258936 | Phr from P. f. | − | − | N.D. | TAACC4) |
| FL13 | pot0846474 | TrmB from T. l. | − | − | trehalose/maltose | ATACT4) |
N.A.: not applicable since these do not have a DBD. N.D.: not determined. N.E.: not examined. P. f.: Pyrococcus furiosus. T. l.: Thermococcus litralis.
pot (Pyrococcus sp. OT3) followed by stop codon positions in the genome (http://www.aist.go.jp/RIODB/archaic).
this study.
Materials and methods
Protein purification
The gene of the FL4 protein from Pyrococcus sp. OT3 was amplified by the polymerase chain reaction (PCR),28) and cloned into the pET28 expression vector. A protein expressed using this vector has a His-tag29) at its N-terminus. The E. coli strain BL21(DE3)/plysE was transformed with the vector, and the gene fl4 was expressed, using an inducer, isopropyl β-D-thiogalactopyranoside (IPTG).
From a culture, 2 l, E. coli cells were collected by centrifugation at 9,000 × g for 10 min at 4 °C, and suspended into 15 ml of PBS buffer (0.4 mM Na2HPO4 and 0.18 mM KH2PO4, adjusted by HCl to pH, 7.4, containing 13.7 mM NaCl and 0.27 mM KCl) containing 1% Triton X100. The supernatant was sonicated twice for 30 sec each, and kept at 75 °C for 10 min, while mixed gently by pipeting. After centrifugation at 27,000 × g for 10 min at 25 °C, 4 M (NH4)2SO4, 30 ml, was added to the super-natant and kept at room temperature for 1 hr. After centrifugation at 9,000 × g for 10 min at 25 °C the sediment was dissolved into 2 ml of buffer A, i.e. 20 mM HEPES (adjusted to pH, 7.4, using KOH) containing 10 mM MgSO4, 1 mM DTT, 1 mM EDTA, 50 mM NaCl, and 5% glycerol, and kept at 85 °C for 10 min. After centrifugation at 9,000 × g for 10 min at 25 °C, the supernatant was dialyzed against 400 ml of buffer A for 2 hrs at room temperature. After centrifugation at 9,000 × g for 10 min at 25 °C, the protein precipitated was dissolved into 1 ml of buffer A containing 1 M urea, and dialyzed against 250 ml of buffer B, 14.3 mM HEPES (adjusted to pH, 7.4, using KOH) containing 7 mM MgSO4, 0.7 mM DTT, 0.7 mM EDTA, 270 mM NaCl, and 27% glycerol, for 2 hrs at room temperature. This process of dialysis was carried out once more overnight. The protein solution was centrifuged at 27,000 × g for 10 min at 25 °C, and filtered through a membrane (the pore size of 0.45 µm, NALGEN, Rochester) to remove large contaminants.
The purified protein formed a single band in an SDS polyacrylamide gel after electrophoresis. Its binding to a column, Ni-NTA spin (QIAGEN, Hilden, Germany), using the His-tag added to its N-terminus, was confirmed.
Preparation of genomic DNA fragments
The genomic DNA molecule of P. OT3 was treated with restriction enzymes, AfaI, AluI, HaeIII, TthHB, HinfI, MseI, Sau3AI, ApaI and MboII, respectively. A mixture of these fragments were subjected to electrophoresis using a gel containing 1.5% SeaPlaque agarose (TaKaRa, Tokyo). From the gel fragments of the size, 100–1,000 bps, were recovered using SUPREC-01 (TaKaRa, Tokyo). Using the DNA blunting kit (TaKaRa, Tokyo), the fragments were ligated to the HincII site of the pBluescript plasmid, pre-treated with bacterial alkaline phosphatase. The plasmid containing various DNA fragments was introduced into an E. coli strain, XL-1-Blue. The E. coli cells were grown on LB plates containing ampicillin, 100 µg/ml, 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal), 130 µg/ml, and IPTG, 1 mM, overnight at 37 °C. A mixture of the plasmid DNAs amplified (see Results) was used as a DNA library in the following round of SELEX experiments.
SELEX Protocol
The DNA library, 5.0 µg of the pBluescript plasmids, FL4, 1.0 µg, and nickel-coated silica beads, 10 µl of a suspension, 250 µl, of materials obtained from a column, Ni-NTA spin (QIAGEN, Hilden, Germany), were mixed into PBS buffer, 100 µl, containing 20% glycerol, 10 mM imidazole, 1.0 µg poly dI/dC, 5 mM β-mercaptoethanol, and varying concentrations of MgSO4, NaCl, and KCl (Table II). The solution was incubated for 15 min at a temperature, 60, 80 or 95 °C. As a negative control, 0.2 µg of the library DNA was used instead of 5.0 µg (see Results).
Table II.
Conditions and efficiencies of SELEX experiments
| DNA (µg) | FL4 (µg) | MgSO4 (mM) | KCl (mM) | NaCl (mM) | °C | No. white | No. blue | Primary W/B* | Secondary W/B** |
|---|---|---|---|---|---|---|---|---|---|
| Optimization 1 | |||||||||
| 5.0 | 1.0 | 5 | 150 | 150 | 60 | 590 | 818 | 0.72 | 1.07 |
| 5.0 | 1.0 | 25 | 150 | 150 | 60 | 563 | 780 | 0.72 | 1.07 |
| 5.0 | 1.0 | 55 | 150 | 150 | 60 | 227 | 208 | 1.09 | 1.63 |
| 5.0 | 1.0 | 105 | 150 | 150 | 60 | 17 | 10 | 1.70 | 2.53 |
| 5.0 | 1.0 | 5 | 350 | 150 | 60 | 483 | 987 | 0.49 | 0.73 |
| 5.0 | 1.0 | 5 | 150 | 350 | 60 | 331 | 548 | 0.60 | 0.89 |
| 0.2 | 1.0 | 5 | 150 | 150 | 60 | 116 | 172 | 0.67 | 1.00 |
|
| |||||||||
| Optimization 2 | |||||||||
| 5.0 | 1.0 | 55 | 150 | 450 | 60 | 299 | 325 | 0.92 | 1.61 |
| 5.0 | 1.0 | 55 | 150 | 550 | 60 | 84 | 56 | 1.50 | 2.63 |
| 5.0 | 1.0 | 55 | 150 | 750 | 60 | 83 | 47 | 1.77 | 3.10 |
| 5.0 | 1.0 | 55 | 150 | 1000 | 60 | 233 | 121 | 1.93 | 3.39 |
| 0.2 | 1.0 | 5 | 150 | 150 | 60 | 236 | 411 | 0.57 | 1.00 |
|
| |||||||||
| Optimization 3 | |||||||||
| 5.0 | 1.0 | 55 | 150 | 450 | 60 | 115 | 224 | 0.51 | 1.65 |
| 5.0 | 1.0 | 55 | 150 | 450 | 80 | 358 | 1068 | 0.34 | 1.10 |
| 5.0 | 1.0 | 55 | 150 | 450 | 95 | 340 | 788 | 0.43 | 1.39 |
| 5.0 | 1.0 | 55 | 150 | 1000 | 60 | 72 | 101 | 0.71 | 2.29 |
| 5.0 | 1.0 | 55 | 150 | 1000 | 80 | 76 | 145 | 0.52 | 1.68 |
| 5.0 | 1.0 | 55 | 150 | 1000 | 95 | 84 | 193 | 0.44 | 1.42 |
| 0.2 | 1.0 | 5 | 150 | 150 | 60 | 188 | 615 | 0.31 | 1.00 |
|
| |||||||||
| 1st round | |||||||||
| 5.0 | 1.0 | 50 | 150 | 1000 | 60 | 320 | 270 | 1.19 | 2.28 |
| 0.2 | 1.0 | 50 | 150 | 1000 | 60 | 317 | 610 | 0.52 | 1.00 |
| 5.0 | 0 | 50 | 150 | 1000 | 60 | 446 | 962 | 0.46 | 0.88 |
|
| |||||||||
| 2nd round | |||||||||
| 5.0 | 1.0 | 50 | 150 | 1000 | 60 | 184 | 247 | 0.74 | 1.34 |
| 0.2 | 1.0 | 50 | 150 | 1000 | 60 | 256 | 466 | 0.55 | 1.00 |
| 5.0 | 0 | 50 | 150 | 1000 | 60 | 1152 | 4328 | 0.27 | 0.49 |
the ratio of No. white to No. blue.
the W/B ratio relative to another W/B observed using 0.2 µg DNA.
After the incubation, silica beads were collected by centrifugation at 800 × g at 25 °C, and, after 500 µl of PBS buffer was added, centrifuged again. This washing process was repeated five times. Then, 200 µl of Tris-EDTA buffer, i.e. 10 mM Tris-HCl buffer (pH = 8.0) containing 0.1 mM EDTA, were added and mixed well. After centrifugation at 800 × g at 25 °C, phenol, 200 µl, was added, and plasmid DNAs were isolated by ethanol precipitation. The plasmid DNAs were suspended into 20 µl of Tris-EDTA buffer. Using 0.5 µl of this plasmid solution E. coli cells XL1-Blue were transformed.
Results
Strategy for optimizing the SELEX protocol
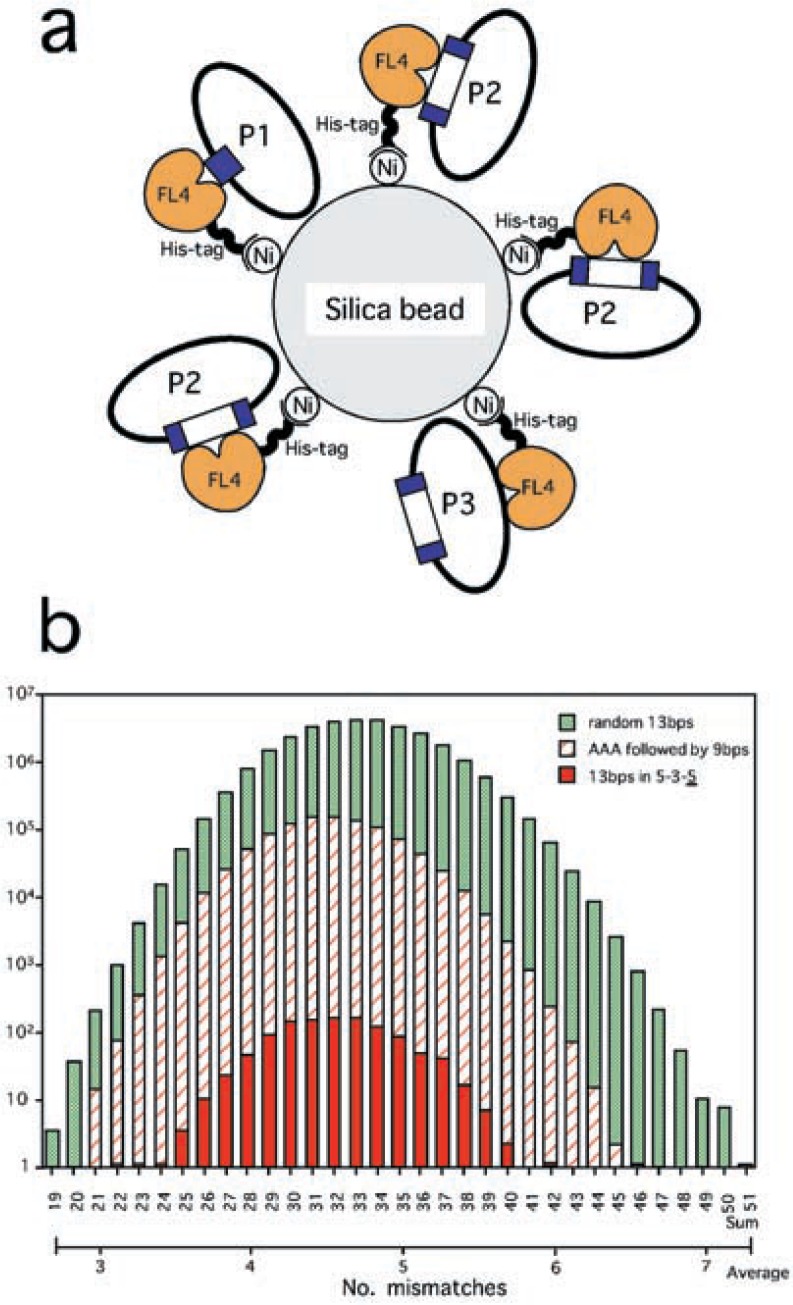
Host E. coli cells transformed with the original pBluescript plasmid are expected to form colonies in a bluish color. This plasmid carries the lacZ gene. Its product, β-galactosidase, catalyzes the substrate X-gal, present in the LB plate, thereby producing this color. When a DNA fragment is inserted into the lacZ gene, β-galactosidase will not be expressed in its original form (Fig. 1a, P2 and P3), yielding the original whitish color of E. coli cells. Thus, the SELEX protocol was optimized, so that the highest ratio of white to blue (W/B) was obtained, and so that the number of white colonies was reasonably high. Here, experiments carried out in the presence of plasmid DNA, 0.2 µg, were considered as negative controls (Table II).
Fig. 1.
The principle of SELEX experiments applied (a), and a histogram of LMNs calculated for the seven fragments selected multiple times by SELEX versus three sets of reference sequences (b). (a) To the surface of silica beads His-tag added FL4 bound through the metal nickel. Plasmids, pBluescript, either containing (P2 and P3) DNA fragments (white boxes) in the lacZ gene (blue edges separated) or not containing (P1), were bound by FL4, thereby selected. However, the site bound by FL4 can be positioned outside the cloning site (P1 and P3), i.e. contamination. The experiments were designed so that the number of type P2 was maximized. (b) A set of randomly combined 13 bps, another set where AAA is followed by randomly combined 9 bps, and a third set of 13 bps in the 5-3-5 arrangement. Ranks are labeled with the sum LMNs as well as the average, i.e. the sum divided by seven.
The SELEX protocol optimized
When the concentration of MgSO4 was changed to 5, 25, 55, and 105 mM, respectively (Table II, Optimization I), in the presence of 150 mM KCl and 150 mM NaCl, the best W/B ratio, 1.70, was obtained with 105 mM MgSO4. However, the absolute number of white colonies, 17, was too small. Thus, the MgSO4 concentration of 55 mM was judged better: the primary W/B ratio was 1.09, and the secondary ratio relative to that found in the negative control was 1.63. When the KCl or NaCl concentration was increased to 350 mM in the presence of 5 mM MgSO4, the W/B ratio was not improved.
In the presence of 55 mM MgSO4, when the NaCl concentration was increased stepwisely to 450, 550, 750, and 1,000 mM, respectively (Table II, Optimization 2), the best primary W/B ratio 1.93 was obtained with 1,000 mM NaCl: the secondary ratio was 3.39. When the experiment was repeated in the presence of 1,000 mM or 450 mM NaCl, and 55 mM MgSO4 at various temperatures, 60–95 °C (Table II, Optimization 3), the best primary W/B ratio, 0.71, and the best secondary ratio, 2.29, were obtained with 1,000 mM NaCl at 60 °C.
On the basis of all these observations, the MgSO4 concentration of 50 mM, the KCl concentration of 150 mM, and the NaCl concentration of 1,000 mM were chosen for the final SELEX protocol at the temperature of 60 °C.
Selection of DNA fragments
In the first round of SELEX experiments (Table II), the primary and secondary W/B ratios observed were 1.19 and 2.28, respectively. In the second round the primary W/B ratio was lower, 1.34, and the secondary ratio was 1.34. Theoretically, the variation of plasmids selected in each round will decrease, thereby concentrating those containing binding-sites of FL4.
After six more rounds, 89 clones were randomly chosen and sequenced. Among them were three copies of the same fragment: FL4-56 (the first entry in Table III). Two copies were found for six other fragments, FL4-2, FL4-25, FL4-26, FL4-29, FL4-40, FL4-74 (Table III, left top). Altogether these 15 copies formed 16.9% of the 89 clones sequenced. The other 74 fragments were of single copies: altogether 81 independent sequences were obtained.
Table III.
Fragments of DNA selected by SELEX
| ID | copy | subfrag. | bps | positions | ID | copy | subfrag. | bps | positions |
|---|---|---|---|---|---|---|---|---|---|
| FL4-56 | 3 | 67 | 1429253–1429319 | FL4-42 | 1 | 198 | 0444403–0444600 | ||
| FL4-2 | 2 | 171 | 0301019–0301189 | FL4-44 | 1 | I | 101 | 0073230–0073330 | |
| FL4-25 | 2 | I | 59 | 0678436–0678494 | II | 133 | 1164592–1164724 | ||
| II | 243 | 0206081–0206323 | FL4-45 | 1 | I | 94 | 0148653–0148746 | ||
| FL4-26 | 2 | I | 230 | 0639360–0639589 | II | 41 | 0206081–0206323 | ||
| II | 56 | 0676358–0676413 | FL4-46 | 1 | 159 | 0346775–0346933 | |||
| III | 37 | 1007989–1008025 | FL4-48 | 1 | 49 | 0681987–0682035 | |||
| FL4-29 | 2 | I | 134 | 1040600–1040733 | FL4-49 | 1 | 170 | 0301019–0301188 | |
| II | 240 | 0662925–0663164 | FL4-50 | 1 | 382 | 0648288–0648669 | |||
| III | 86 | 0900779–0900864 | FL4-51 | 1 | 91 | 1466318–1466408 | |||
| FL4-40 | 2 | 210 | 1347230–1347439 | FL4-52 | 1 | 119 | 0524465–0524583 | ||
| FL4-74 | 2 | 389 | 1604477–1604865 | FL4-53 | 1 | 89 | 0793036–0793124 | ||
| FL4-1 | 1 | 142 | 1567873–1568014 | FL4-54 | 1 | 105 | 1540946–1541050 | ||
| FL4-2 | 1 | 170 | 0301019–0301188 | FL4-55 | 1 | 242 | 1515503–1515744 | ||
| FL4-3 | 1 | 42 | 0715037–0715078 | FL4-58 | 1 | 110 | 0075175–0075284 | ||
| FL4-5 | 1 | I | 107 | 1442314–1442420 | FL4-59 | 1 | 31 | 1017813–1017843 | |
| II | 242 | 0476169–0476410 | FL4-60 | 1 | I | 54 | 1287506–1287559 | ||
| III | 73 | 1231109–1231181 | II | 663 | E. coli K12 | ||||
| FL4-6 | 1 | 217 | 0292616–0292832 | FL4-61 | 1 | 130 | 1075739–1075868 | ||
| FL4-8 | 1 | 160 | 1165979–1166138 | FL4-62 | 1 | 110 | 1149647–1149756 | ||
| FL4-9 | 1 | I | 34 | 0729947–0729980 | FL4-63 | 1 | 86 | 0799661–0797746 | |
| II | 93 | no homology | FL4-64 | 1 | 80 | 1287923–1288002 | |||
| FL4-10 | 1 | I | 62 | 0214930–0214991 | FL4-65 | 1 | 274 | 1660946–1661219 | |
| II | 31 | 0022141–0022171 | FL4-66 | 1 | I | 249 | 0709338–0709586 | ||
| FL4-11 | 1 | 272 | 0048106–0048377 | II | 202 | 0830728–0830929 | |||
| FL4-12 | 1 | 39 | 1617471–1617509 | FL4-68 | 1 | 37 | 1027108–1027144 | ||
| FL4-13 | 1 | 65 | 1728886–1728940 | FL4-69 | 1 | 94 | 1539656–1539749 | ||
| FL4-16 | 1 | 114 | 1670785–1670898 | FL4-70 | 1 | 52 | 1381522–1381573 | ||
| FL4-17 | 1 | 371 | 1235147–1235517 | FL4-72 | 1 | I | 126 | 0139160–0139285 | |
| FL4-18 | 1 | 102 | 0754580–0754681 | II | 137 | 0019022–0019158 | |||
| FL4-19 | 1 | I | 244 | 0009691–0009934 | FL4-75 | 1 | 210 | 1347230–1347439 | |
| II | 165 | 1042318–1042482 | FL4-76 | 1 | 214 | 1507767–1507980 | |||
| FL4-20 | 1 | 304 | 1060276–1060579 | FL4-77 | 1 | I | 153 | 1341065–1341217 | |
| FL4-21 | 1 | 109 | 0087812–0087920 | II | 237 | 1731960–1732196 | |||
| FL4-22 | 1 | 80 | 0481286–0481365 | FL4-79 | 1 | 198 | 0052204–0052401 | ||
| FL4-23 | 1 | 50 | 0927463–0927512 | FL4-80 | 1 | 64 | 0526230–0526293 | ||
| FL4-24 | 1 | 46 | 1588968–1589013 | FL4-81 | 1 | I | 109 | 1507649–1507757 | |
| FL4-27 | 1 | I | 70 | 0888443–0888512 | II | 115 | 1695509–1695623 | ||
| II | 111 | 1381434–1391544 | FL4-82 | 1 | 412 | 1506389–1506800 | |||
| FL4-28 | 1 | I | 34 | 0792065–0792198 | FL4-83 | 1 | 118 | 0189560–0189677 | |
| II | 91 | 0637676–0637766 | FL4-84 | 1 | 49 | 1467382–1467430 | |||
| FL4-30 | 1 | 45 | 1713156–1713200 | FL4-86 | 1 | 110 | 0348509–0348618 | ||
| FL4-31 | 1 | 168 | 1140474–1140641 | FL4-88 | 1 | 219 | 1240256–1240474 | ||
| FL4-32 | 1 | 28 | 0375531–0375558 | FL4-91 | 1 | 152 | 0212718–0212869 | ||
| FL4-33 | 1 | 52 | 0072318–0072369 | FL4-92 | 1 | 358 | 0313462–0313819 | ||
| FL4-34 | 1 | I | 103 | 0026339–0026441 | FL4-93 | 1 | 114 | 1672618–1672731 | |
| II | 90 | 0481727–0481816 | FL4-94 | 1 | 69 | 0322370–0322438 | |||
| FL4-35 | 1 | 34 | 1658671–1658704 | FL4-95 | 1 | I | 187 | 0962153–0962339 | |
| FL4-36 | 1 | 211 | 1273014–1273224 | II | 197 | 0142757–0142953 | |||
| FL4-37 | 1 | 53 | 0777830–0777882 | III | 118 | 0657477–0659594 | |||
| FL4-38 | 1 | 131 | 1471475–1471605 | IV | 76 | 0129288–0129363 | |||
| FL4-41 | 1 | 79 | 1681573–1681651 | FL4-96 | 1 | 270 | 0771434–0771703 |
From another point of view, 14 of the 81 fragments were found as containing two independent subfragments each (subfragments I and II in Table III), 3 fragments (FL4-5, 26, 29) as containing three subfragments each, and yet another fragment (FL4-95) as containing four subfragments: altogether these forming 22.2% of the 81 fragments. With including fragments having single subfragments only, the average number of subfragments found in the 81 fragments was 1.28. Of all the 104 different subfragments, FL4-60-II was found originating in E. coli but not in P. OT3: a contamination. Another subfragment, FL4-9-I, was also a contamination but from an unknown origin. The average length of the subfragments was 185 bps. The average G: C content of the subfragments was 42%, which is the same as that of the genome of P. OT3.
In what follows, the seven fragments selected multiple times, FL4-2, 25, 26, 29, 40, 56, 74, are further analyzed, since the possibility of their containing real binding sites is higher than that of other fragments selected only single time.
Discussion and analysis
Thirteen basepairs shared by the seven fragments with the minimum mismatches
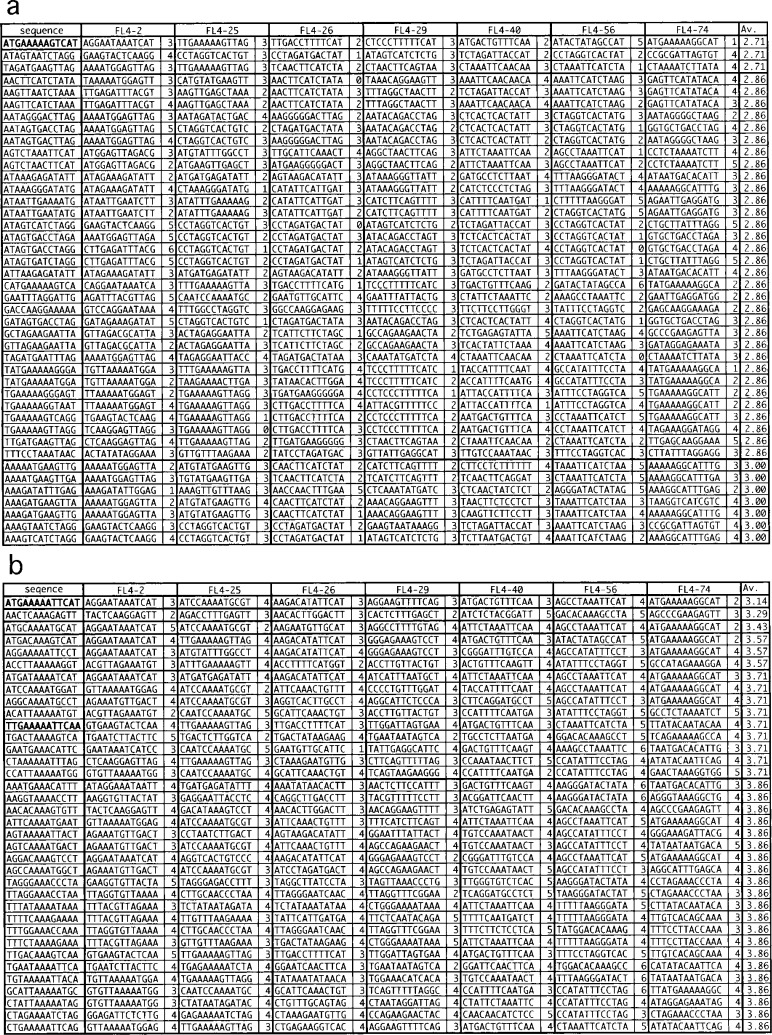
Any DNA-binding domain (DBD) can cover only one side of DNA for ∼5 bps, and two such DBDs in a dimer are often separated by ∼10 bps or shorter along the DNA. Thus the sequence recognized by a dimer of a transcription factor will not much exceed ∼15 bps. Indeed, dimmers of FFRPs recognize 13 bps in the 5-3-5 arrangement (see Introduction). Thus, for each 13 bps randomly combined, the number of mismatches found with each of the seven fragments at its best resembling part was calculated: the least mismatch number, LMN (Fig. 2a).
Fig. 2.
Thirteen the best conserved among the seven DNA fragments selected multiple times by SELEX (a), and a selection of those in the 5-3-5 arrangements (b). The least mismatch numbers (LMNs) found with each fragment and their average are shown for each reference sequence.
One of the three random 13 bps best conserved among the seven fragments selected multiple times was ATGAAAAAGTCAT, with the average LMN, 2.71 (Fig. 2a, highlighted in bold). This sequence is closely related with a 5-3-5 sequence, ATGAAAAATTCAT, having only one mismatch: here bases the same as in ATGAAAAAGTCAT are underlined. In fact, ATGAA[AAA/TTT]TTCAT was found to be the single best 5-3-5 sequence conserved among the seven fragments with the average LMN of 3.14 (Fig. 2b, highlighted in bold).
Another 5-3-5 sequence, which needs to be considered, is TTGAA[AAA/TTT]TTCAA. Many transcription factors auto-regulate the genes coding themselves, and so might be FL4. Upstream of the fl4 gene, the sequence TTGAAATTTACAA is positioned between a putative TATA box and an SD signal (Fig. 3). This is a typical formation for an FFRP to act as a repressor.1),9) The sequence resembles TTGAATTTTTCAA more than ATGAATTTTTCAT. The average LMN of 3.71 was calculated between TTGAATTTTTCAA and the seven selected fragments (Fig. 2b).
Fig. 3.
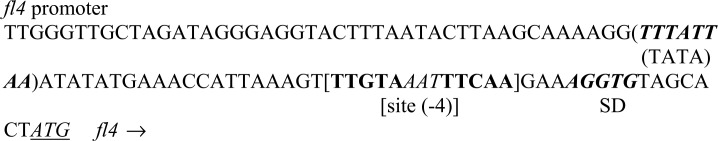
The nucleotide sequence of the region upstream of the fl4 gene in the P. OT3 genome. Candidates for a TATA box, an SD signal and the start codon are indicated. A putative FL4 binding site is also indicated with the number of mismatches with the sequence ATGAA[AAA/TTT]TTCAT. The arrow shows the direction of transcription.
Possible repression of fl9 gene by FL4 protein
The average LMN found between ATGAA[AAA/TTT]TTCAT and the seven fragments was 3.14, which is better than our empirical threshold for theoretical identification of sites bound by FFRPs, ∼4. Yet FL4-25 and FL4-56 are on the border (Fig. 2b). Not all the seven fragments might contain sites functioning as real signal sequences. On the other hand, even when the score is below 4, the site might function as a signal sequence, when another binding site, even if it is less ideal, is positioned nearby: a cooperative interaction. When 7–8 bps are inserted between a pair of 13 bps, the two sites repeat with a periodicity of 20–21 bps, i.e. two full helical turns of DNA. With this arrangement, a pair of dimers can contact each other on the same side of the DNA, thereby forming a tetramer.22) More generally, the number of basepairs expected to be inserted is ∼[10–11] × N, where N is an integer.
In FL4-56, and the part immediately downstream (Fig. 4f, shown by characters in lower case), three putative binding sites, two with four mismatches with ATGAA[AAA/TTT]TTCAT, and the other with five, were found as repeating with insertions of 18 and 20, respectively, positioned upstream of the gene pot1428536. The second and third sites were found as sandwiching a TATA-box, CTTAAAAA (Fig. 4f): a formation of repressing transcription of the gene. In FL4-2 (Fig. 4a), three putative binding sites, with three to five mismatches with ATGAA[AAA/TTT]TTCAT, were found repeating with insertions of 17 bps and 28 bps, respectively. The third site overlaps onto a TATA box, ATTGAATC, positioned upstream of gene pot0301583. This arrangement also represents a repression mode.
Fig. 4.
The nucleotide sequences of the seven fragments selected multiple times by SELEX experiments. Candidates for TATA boxes, SD signals and the start codons, ATG and TTG, are indicated. Putative FL4-binding sites are also indicated with the numbers of mismatches with the sequence ATGAA[AAA/TTT]TTCAT. Gene-coding regions are underlined. Except for (c), (e) and (g), possible modes of regulation, i.e. repression or activation, are written. In (c), (e) and (g) putative binding sites are found inside gene coding regions only, although still it is possible that they are designed for repression by FL4. In (b) two genes, pot0206233 and pot0206777, are overlapping onto each other inside an operon, and the 3′-end of the former and the 5′-end of the latter are indicated by parentheses,)) and (, respectively. Nucleotides found immediately outside the fragments are shown in lower case in (d), (f) and (g). For FL4-25 and 36 containing multiple subfragments, only single subfragments each are shown, since the other subfragments do not contain 13 bps closely related with ATGAA[AAA/TTT]TTCAT. For FL4-29, all the three sub-fragments are shown.
Importantly, gene pot0301583 codes for another FFRP, FL9 (see Table I for the full ID). Immediately downstream of the fl9 gene, another gene codes DM2 in the same direction, most likely forming an operon. The protein DM2 is one of the three demi-FFRPs20) coded in the genome of P. OT3, having assembly domains only of full length FFRPs, e.g. FL4 and FL9. The two proteins, DM2 and FL9, are able to interact (Makino, K. et al., unpublished). These facts hint at the presence of a transcription network organized by FFRPs.
Possible transcription activation of pot1040906 by FL4.
Fragment FL4-29 was a chimera of three sub-fragments (Table III). It is not known which one of the three contains a binding site of FL4. In subfragment III the region upstream of gene pot1040906, coding a protein of an unknown function, was cloned (Fig. 4d), but the two other subfragments contain gene-coding regions only. In subfragment III two sites with five mismatches each with ATGAA[AAA/TTT]TTCAT were positioned with an insertion of 36 bps, which is not so different from 7 plus 10 multiplied by 3. Further upstream of the two sites, another site with three mismatches was positioned with an insertion of 9 bps, although not whole of this site was included in FL4-29III (shown in lower case in Fig. 4d). Downstream of the third site, separated by 3 bps, a putative TATA box is found. This particular arrangement fits well into the pattern of those predicted for activating transcription of genes by FFRPs,1),5),9) this time that of pot1040906.
For the two other chimeric fragments, FL4-25 and 36, only single subfragments each are shown in Fig. 4, since the other subfragments do not contain 13 bps closely related with ATGAA[AAA/TTT]TTCAT.
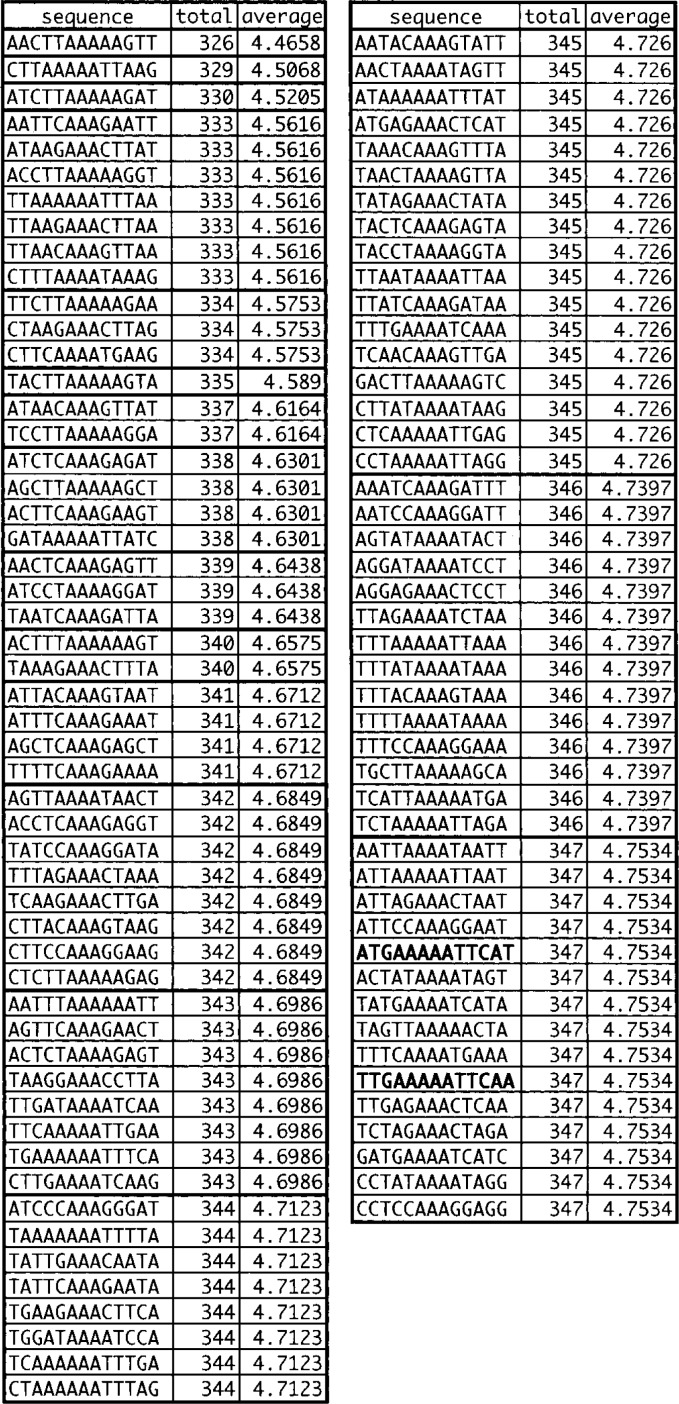
Other statistical analyses
When LMNs were calculated between each of the 1,024 (i.e. 45) sequences in the 5-3-5 arrangement and the 73 fragments selected single time only (Fig. 5), the sequences, ATGAA[AAA/TTT]TTCAT and TTGAA[AAA/TTT]TTCAA, were given scores inside top 10%. The sum LMN was 347 and the average LMN was 4.75 with both 5-3-5 sequences. These observations are consistent with the idea that the 74 fragments are a mixture of those containing real binding sites, and a larger number of contaminants.
Fig. 5.
Thirteen bases the best conserved among 37 fragments selected single time only by SELEX. The sum and average least mismatch numbers (LMNs) found with the fragments are shown for each reference sequence.
The average of average LMNs calculated between a set of randomly combined 13 bps and the seven fragments was 5.00 (Fig. 1b). When reference sequences were restricted to those having AAA at the 5′ ends, or TTT at the 3′ ends, the secondary average of LMNs was improved to 4.79. This was because of the A:T content in the genome, 58%, which was higher than the average A:T of 50% in the first set and closer to that in the second set, 63%. The same A:T% was kept in a third set of 13 bps in the 5-3-5 arrangement. With this set, the average LMN was further improved to 4.51, suggesting that real-binding sites did have this type of arrangement.
The scores calculated with the second set distributed more or less symmetrically to the two ends, but those calculated with the third set tailed more to the minimal mismatching end (Fig. 1b). The presence of a small number of 5-3-5 sequences having the smallest LMNs, i.e. ATGAA[AAA/TTT]TTCAT and its close variants, ATGCA[AAA/TTT]TGCAT and AGGAA[AAA/TTT]TTCCT, suggests that these sequences are indeed related with real binding sites of FL4.
Acknowledgments
This work was supported by CREST (Core Research for Evolutionary Science and Technology) program of JST (Japan Science and Technology Agency) in the research area PSFM (Protein Structure and Functional Mechanisms). We thank Dr. Y. Azuma for his help at an early stage of this study.
References
- 1.Yokoyama, K., Ishijima, S. A., Clowney, L., Koike, H., Aramaki, H., Tanaka, C., Makino, K., and Suzuki, M. (2005) FEMS Microbiol. Rev. (in press). [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama, K., Ihara, M., Ebihara, S., and Suzuki, M. (2005) Proc. Jpn. Acad., Ser. B 81, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki, M. (2005) Proc. Jpn. Acad., Ser. B 81, 403–409. [Google Scholar]
- 4.Suzuki, M. (2005) Proc. Jpn. Acad., Ser. B 81, 334–348. [Google Scholar]
- 5.Koike, H., Yokoyama, K., Kawashima, T., Yamasaki, T., Makino, S., Clowney, L., and Suzuki, M. (2005) Proc. Jpn. Acad., Ser. B 81, 278–290. [Google Scholar]
- 6.Yokoyama, K., Ebihara, S., Kikuchi, T., and Suzuki, M. (2005) Proc. Jpn. Acad., Ser. B 81, 64–75. [Google Scholar]
- 7.Sakuma, M., Nakamura, M., Koike, H., and Suzuki, M. (2005) Proc. Jpn. Acad., Ser. B 81, 110–116. [Google Scholar]
- 8.Sakuma, M., Koike, H., and Suzuki, M. (2005) Proc. Jpn. Acad., Ser. B 81, 26–32. [Google Scholar]
- 9.Yokoyama, K., and Suzuki, M. (2005) Proc. Jpn. Acad., Ser. B 81, 129–139. [Google Scholar]
- 10.Kawashima, T., Yokoyama, K., Higuchi, S., and Suzuki, M. (2005) Proc. Jpn. Acad., Ser. B 81, 204–219. [Google Scholar]
- 11.Koike, H., Ishijima, S. A., Clowney, L., and Suzuki, M. (2004) Proc. Natl. Acad. Sci. USA 101, 2840–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishijima, S. A., Clowney, L., Koike, H., and Suzuki, M. (2004) Proc. Jpn. Acad., Ser. B 80, 22–27. [Google Scholar]
- 13.Ishijima, S. A., Clowney, L., Koike, H., and Suzuki, M. (2004) Proc. Jpn. Acad., Ser. B 80, 107–113. [Google Scholar]
- 14.Clowney, L., Ishijima, S. A., and Suzuki, M. (2004) Proc. Jpn. Acad., Ser. B 80, 148–155. [Google Scholar]
- 15.Ishijima, S. A., Clowney, L., and Suzuki, M. (2004) Proc. Jpn. Acad., Ser. B 80, 183–188. [Google Scholar]
- 16.Ishijima, S. A., Clowney, L., and Suzuki, M. (2004) Proc. Jpn. Acad., Ser. B 80, 236–243. [Google Scholar]
- 17.Ishijima, S. A., Clowney, L., and Suzuki, M. (2004) Proc. Jpn. Acad., Ser. B 80, 459–468. [Google Scholar]
- 18.Suzuki, M. (2003) Proc. Jpn. Acad., Ser. B 79, 274–289. [Google Scholar]
- 19.Koike, H., Sakuma, M., Mikami, A., Amamo, N., and Suzuki, M. (2003) Proc. Jpn. Acad., Ser. B 79, 63–69. [Google Scholar]
- 20.Suzuki, M., Amano, N., and Koike, H. (2003) Proc. Jpn. Acad., Ser. B 79, 92–98. [Google Scholar]
- 21.Suzuki, M., and Koike, H. (2003) Proc. Jpn. Acad., Ser. B 79, 114–119. [Google Scholar]
- 22.Suzuki, M. (2003) Proc. Jpn. Acad., Ser. B 79, 213–222. [Google Scholar]
- 23.Suzuki, M., Aramaki, H., and Koike, H. (2003) Proc. Jpn. Acad., Ser. B 79, 242–247. [Google Scholar]
- 24.Ishijima, S. A., Clowney, L., Koike, H., and Suzuki, M. (2003) Proc. Jpn. Acad., Ser. B 79, 299–304. [Google Scholar]
- 25.Kudo, N., Allen, M. D., Koike, H., Katsuya, Y., and Suzuki, M. (2001) Acta Cryst. D57, 469–471. [DOI] [PubMed] [Google Scholar]
- 26.Tuerk, C., and Gold, L. (1990) Science 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 27.Kawarabayasi, Y., Sawada, M., Horikawa, H., Haikawa, Y, Hino, Y., Yamamoto, S., Sekine, M., Baba, S., Kosugi, H., Hosoyama, A.et al. (1998) DNA Res. 5, 55–76. [DOI] [PubMed] [Google Scholar]
- 28.Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn, G. T., Mullis, K. B., and Erlich, H. A. (1988) Science 239, 487–491. [DOI] [PubMed] [Google Scholar]
- 29.Hochuli, E., Dobeli, H., and Schacher, A. (1987) J. Chromatogr. A411, 177–184. [DOI] [PubMed] [Google Scholar]