Figure 1.
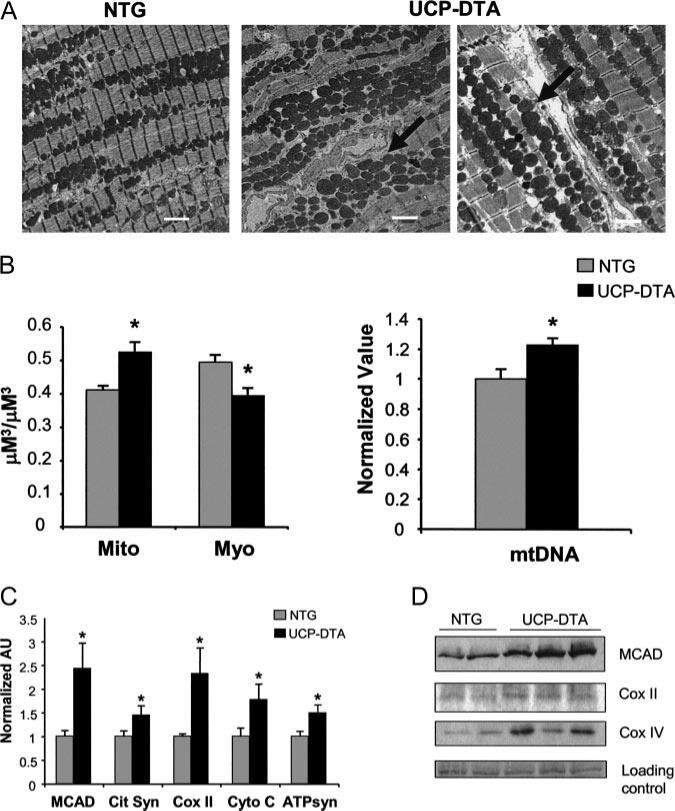
Mitochondrial biogenesis in insulin-resistant hearts. A, Representative electron micrographs of papillary muscles from NTG and insulin-resistant UCP-DTA hearts. White bar=2 μm. Arrows indicate areas of increased mitochondria. B, Quantitative morphometric measurement (left) of mitochondrial (Mito) or myofibril (Myo) cellular volume density (μm3/μm3) based on analysis of electron micrographs (n=6 animals/group). Mean cardiac mtDNA levels (right) determined by real-time PCR analysis shown as arbitrary units (AU; n=5 to 7 hearts/group) normalized to the NTG value (=1.0). C, Results of real-time rtPCR analysis of mitochondrial metabolic genes in insulin-resistant UCP-DTA and NTG control hearts. Cit Syn indicates citrate synthase; Cyto C, cytochrome c; and ATPsyn, ATP synthase-β; other abbreviations defined in text. n=6 to 9 mice/group. Bars represent mean (±SE) AU normalized to the corresponding NTG value. *P<0.05 compared with NTG control. D, Representative immunoblots performed with protein prepared from NTG and insulin-resistant UCP-DTA hearts with medium-chain acyl-CoA dehydrogenase, COX II, and COX IV antibodies. Representative ponceau-stained bands are shown as a loading control. MCAD indicates medium-chain acyl-CoA dehydrogenase.