Figure 3. Screening of candidate target genes of miR-638 in CRC.
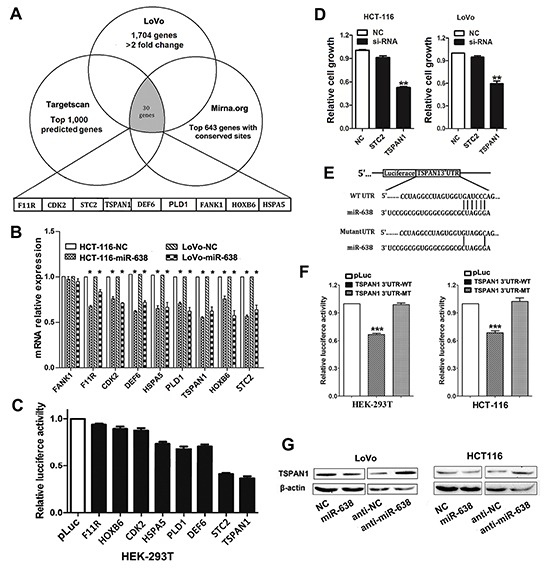
(A) Initial screening of miR-638 target genes in LoVo cells transfected with miR-638 mimic using microarray and bioinformatic predictions. A total of nine downregulated genes were selected from the 30 genes in the initial screening basing on the functional analysis. (B) Validation of the microarray results in both HCT-116 and LoVo cells by quantitative reverse transcription polymerase chain reaction (qRT-PCR). A total of eight genes were indeed downregulated in the miR-638-transfected CRC cells compared to the control cells (*p<0.05). (C) Two candidate genes (STC2 and TSPAN1) were selected for their significantly decreased luciferase activity in the miR-638-transfected HEK-293T cells. (D) Proliferation assays performed using CRC cell lines (HCT-116 and LoVo) transfected with siRNA directed against two candidate genes. Depleted TSPAN1 expression showed the most obvious growth repression effect. ** p<0.01 compared to NC. (E) Schematic of the wild-type or mutant 3′UTRs of the TSPAN1 vector constructs. The complementary site of the seed region of miR-638 was selected for mutation. (F) The relative luciferase activity assays of luciferase reporters with the TSPAN1 wild-type (WT) or mutant-type (MT) 3′UTR were performed in HEK-293T and HCT-116 cells after cotransfection with miR-638 mimic. Luciferase activity was determined at 24 h after transfection and normalized to the Renilla luciferase activity. The luciferase activity generated by pLuc in each experiment was set as 1(***p<0.001 compared to the pLuc control). (G) The protein levels of TSPAN1 were determined by western blot assays using LoVo and HCT-116 cells transfected with miR-638 mimic, miR-638 inhibitor (anti-miR-638) or the corresponding negative control (NC or anti-NC). Beta-actin protein served as an internal control.
