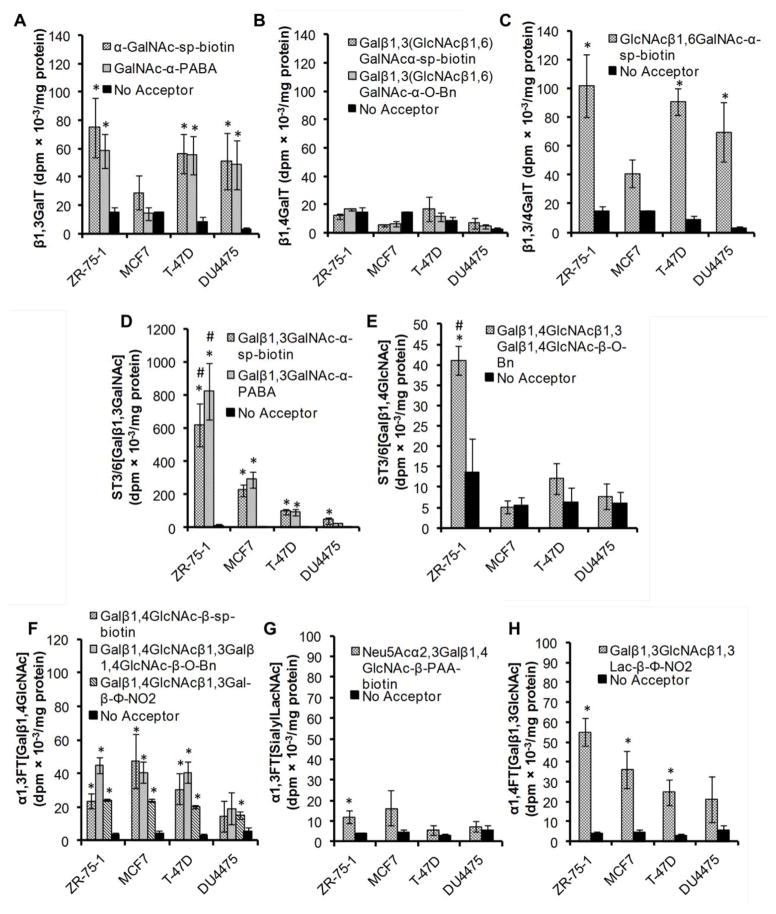
Figure 2. Enzyme activity in breast cancer cell lines.
Product formed was quantified in terms of dpm/mg of protein in cell lysate for four breast cancer cell lines. A. β1,3GalT activity was measured using acceptors with GalNAcα reactive groups. B. β1,4GalT was quantified using acceptors with Galβ1,3(GlcNAcβ1,6)GalNAcα unit. C. β1,3/4GalT activity was measured using GlcNAcβ1,6GalNAcα-sp-biotin, a substrate that can simultaneously measure both enzyme activities. D. ST3/6[Galβ1,3GalNAc] activity was measured using acceptors with the Galβ1,3GalNAc unit. E. ST3/6[Galβ1,4GlcNAc] was quantified using acceptors with LacNAc [Galβ1,4GlcNAcβ] unit. F. α1,3FT[Galβ1,4GlcNAc] activity measured using acceptors with LacNAc unit. G.α1,3FT[sialylLacNAc] was quantified using Neu5Acα2,3Galβ1,4GlcNAcβ-PAA-biotin. Previous studies show thatα1,2FT activity is low in breast cancer cells. H. α1,4FT[Galβ1,3GlcNAc] activity was measured using the acceptor Galβ1,3GlcNAcβ1,3lacβ-Φ-NO2. *indicates P<0.05 with respect to the “No Acceptor” control in the respective cell line. #indicates P<0.05 with respect to all other cell lines.