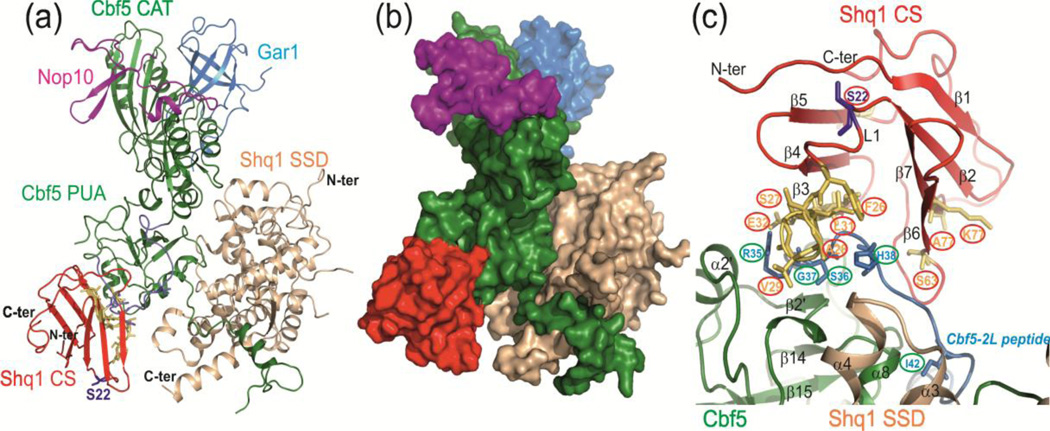
Fig. 4.
NMR CSP data-driven HADDOCK model structure of yCS bound to yeast Cbf5-Nop10-Gar1-Shq1 SSD complex. (a) Model with lowest HADDOCK score with proteins shown as ribbons. The model is based on the structures of yeast Cbf5-Nop10-Gar1-Shq1 SSD complex (PDB 3UAI) and Shq1 yCS (PDB 3EUD). Cbf5 is green, Nop10 is magenta, Gar1 is cyan, Shq1 SSD is wheat, and Shq1 CS is red. The yCS surface identified to interact with Cbf5 peptide using NMR CSP is shown in yellow, and the Cbf5-2S/L peptide position is shown in blue. (b) Surface view of the model, showing the ‘capture’ of Cbf5 by Shq1 CS and SSD. (c) Close up view of interactions of yCS with the Cbf5 PUA domain. Positions of key residues used to drive modeling are shown.