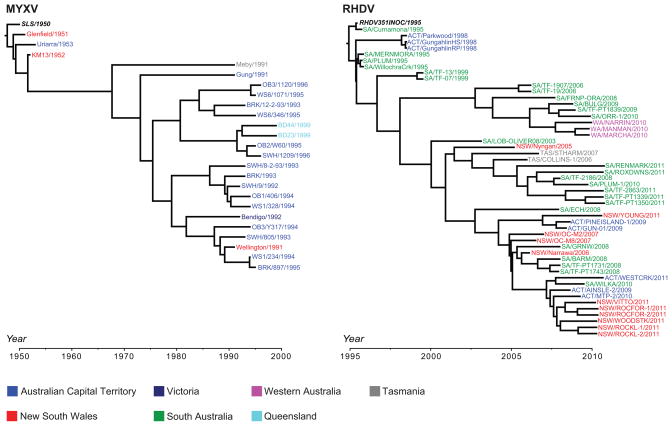
Figure 3. Geographic structure of MYXV and RHDV in Australia.
Time-scaled phylogenetic trees were estimated using the Bayesian Markov chain Monte Carlo method available in the BEAST package [81]. Both MYXV and RHDV analyses utilized the GTR+Γ model of nucleotide substitution, a relaxed molecular clock, and a constant population size coalescent prior. The MYXV data set comprised 25 complete genome sequences (alignment of 163,555 nt), while the RHDV data set comprised 51 capsid (VP60) sequences of 1,737 nt. The sequence names are colored according to the Australian state/territory from where they were isolated. The sequences in bold italics are the strains originally released in 1950 (MYXV) and 1995 (RHDV).