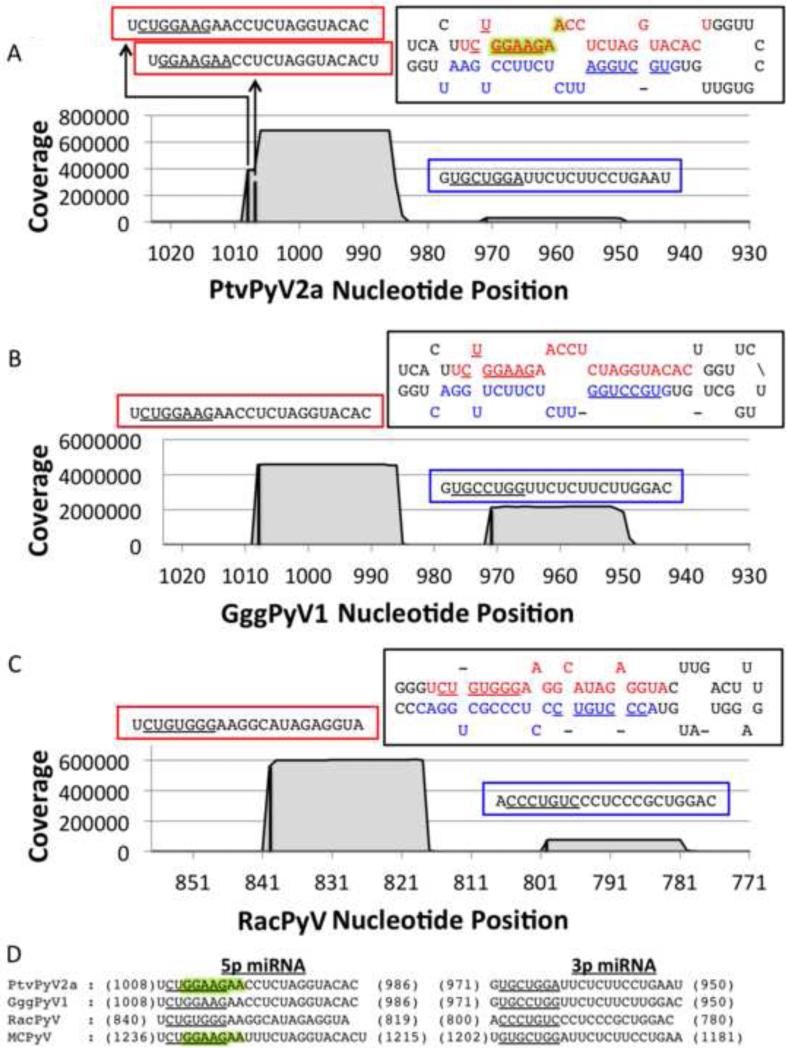
Fig. 2. The PtvPyV2a, GggPyV1 and RacPyV miRNAs are partially conserved.
(A - C) Coverage plot of the small RNA deep sequencing reads from (A) PtvPyV2a, (B) GggPyV1 and (C) RacPyV miRNA expression-vector-transfected 293T cells. The number of reads (y axis) was mapped onto the corresponding PyV genome (x axis). The black bars indicate the start position of each miRNA and the coverage is represented by the grey filled area. All three PyVs produce dominant 5p derivative miRNAs (indicated in the red boxes) and less abundant 3p derivative miRNAs (indicated in the blue boxes). Each miRNA sequence is indicated above its corresponding peak, with the seed sequences (nucleotide positions 2 to 8) underlined. The 5p (red) and 3p (blue) derivative miRNAs are indicated in each hairpin structure, with their seed sequence underlined. PtvPyV2a expresses two different species of the 5p derivative miRNAs. The seed sequences of the major and minor species are underlined and highlighted in green, respectively, in the hairpin structure. (D) The PtvPyV2a, GggPyV1 and RacPyV miRNAs share partial sequence identity with the MCPyV miRNAs. Sequence alignment of the 5p and 3p derivative miRNAs from PtvPyV2a, GggPyV1a and RacPyV and MCPyV miRNA sequences (16, 47). The 5’ and 3’ coordinates of each derivative miRNA are indicated and the seed sequence for each miRNA is underlined. The seed sequences from the major and minor form of the PtvPyV2a 5p derivative miRNAs are underlined and highlighted in green respectively. The seed sequence from the MCPyV 5p derivative miRNA reported by Lee et al. and Seo et al. (16, 47) are underlined and highlighted in green respectively.