Abstract
Phosphodiesterases-6 (PDE6) are essential effector enzymes in vertebrate photoreceptor cells. Mutations in rod and cone PDE6 cause recessive retinitis pigmentosa and achromatopsia, respectively. The mechanisms of missense PDE6 mutations underlying severe visual disorders are poorly understood. To probe these mechanisms, we expressed seven known missense mutants of cone PDE6C in rods of transgenic X. laevis and examined their stability and compartmentalization. PDE6C proteins with mutations in the catalytic domain, H602L and E790K, displayed modestly reduced proteolytic stability, but they were properly targeted to the outer segment of photoreceptor cells. Mutations in the regulatory GAF domains, R104W, Y323N, and P391L led to a proteolytic degradation of the proteins involving a cleavage in the GAFb domain. Lastly, the R29W and M455V mutations residing outside the conserved PDE6 domains produced a pattern of subcellular compartmentalization different from that of PDE6C. Thus, our results suggest a spectrum of mechanisms of missense PDE6C mutations in achromatopsia including catalytic defects, protein mislocalization, or a specific sequence of proteolytic degradation.
1. Introduction
Cyclic-nucleotide phosphodiesterases (PDE) of the sixth family (PDE6) are the key effector enzymes in phototransduction in rods and cones (Fu and Yau, 2007, Arshavsky and Burns, 2012). The enzyme catalytic core is a heterodimer of PDE6A and PDE6B subunits in rod PDE6 and a homodimer of PDE6C subunits in cone PDE6. Mutations in the PDE6A and PDE6B genes are responsible for a significant fraction of recessive retinitis pigmentosa (RP), an inherited degenerative retina disease leading to blindness (McLaughlin et al., 1995, Dryja et al., 1999). Mutations in PDE6C cause autosomal recessive achromatopsia (ACHM) (Chang et al., 2009, Thiadens et al., 2009, Grau et al., 2011). ACHM results from a loss of cone function and is characterized by low visual acuity and lack of colour discrimination. Many mutations in PDE6, including nonsense mutations, splice defects, and frame shifts are certain to cause loss of expression, misfolding, and/or loss of PDE6 function. However, the mechanisms of missense PDE6 mutations in rods and cones leading to retina disease are largely unknown.
Missense mutations may alter PDE6 function or interfere with transport of functional PDE6 to the outer segment (OS), a specialized ciliary compartment of photoreceptor cells. Proper transport of PDE6 in photoreceptors is critically important for the function and survival of rods and cones. Lack of functional PDE6 in the rod OS leads to elevation of cGMP levels and causes rapid retinal degeneration (RD) in animal models and humans (Farber and Lolley, 1974, Bowes et al., 1990, Pittler and Baehr, 1991, Liu et al., 2004, Ramamurthy et al., 2004). The possibility that abnormal PDE6 trafficking may underlie RP is highlighted by the PDE6B mutation L854V in the protein C-terminal CAAX motif (Veske et al., 1995). Isoprenylation modifications of PDE6 are critical to the PDE6 interaction with membranes and transport (Anant et al., 1992, Catty et al., 1992, Veske et al., 1995, Karan et al., 2008). The C-terminal CAAX boxes of mammalian PDE6A and PDE6B specify farnesylation and geranylgeranylation, respectively (Anant et al., 1992). In an attempt to characterize PDE6C mutants linked to ACHM, several missense mutations were previously introduced into a chimeric PDE5/PDE6C enzyme expressed in sf9 cells (Grau et al., 2011). These mutants indicated either a loss or reduction of the catalytic activity (Grau et al., 2011). However, the use of PDE5/PDE6C chimeras to study effects of PDE6 mutations has severe limitations. PDE5/PDE6C chimeras contained the catalytic domain of PDE5 and thus, they are essentially PDE5-like enzymes. Furthermore, the process of PDE6 folding, assembly, and trafficking in rod and cones is complex and it involves photoreceptor-specific protein machinery (Ramamurthy et al., 2004, Karan et al., 2008). These aspects cannot be recapitulated using the chimera/insect cell system.
Our previous studies demonstrated that transgenic X. laevis is a robust system to investigate PDE6 and its transport (Muradov et al., 2009, Muradov et al., 2010). Human EGFP-tagged PDE6C ectopically expressed in rods of transgenic X. laevis traffics correctly to the OS. In the OS, EGFP-PDE6C concentrates at the disc rims and co-localizes with endogenous frog rod PDE6 (Muradov et al., 2009). Recently, using transgenic X. laevis we demonstrated that the GAFa domain of PDE6 contains an OS localization signal (Cheguru et al., 2014). Here, we expressed seven known ACHM-linked missense mutants of PDE6C in rods of X. laevis to examine the mutations’ effects on the protein expression, stability, and transport. The PDE6C mutants demonstrated distinct patterns of protein stability and compartmentalization depending on the domain(s) harboring the mutation and indicated a spectrum of mechanisms in achromatopsia.
2. Materials and methods
2.1. Generation of PDE6C mutants for expression in transgenic Xenopus rods
The pXOP-EGFP-PDE6C vector for expression of the EGFP-fused human cone PDE6 in transgenic X. laevis was used as a template for introduction of the following mutations: R29W, R104W, Y323N, P391L, M455V, H602L, and E790K. Mutations were generated using QuikChange mutagenesis (Agilent), or a two-step PCR-directed mutagenesis as described previously (Natochin et al., 1998). Briefly, in the first round PCR, mutant primers were paired with a primer carrying a unique restriction site. PCR products after the first round were purified on agarose gel and used as primers for a second round PCR amplification in combination with a primer containing another restriction site. The final PCR-products were subcloned in the pXOP-EGFP-PDE6C vector using selected restriction sites. In addition, the QuikChange protocol was used to mutate sequence 829EAKKQE834 to EYMPTE thereby creating an epitope for the monoclonal antibody EE within the C-terminal region of PDE6C. All constructs were confirmed by automated DNA sequencing at the Iowa Institute of Human Genetics Genomics Division. DNA was purified using a Qiagen Midiprep kit, digested with XhoI to linearize the plasmids, and re-purified with Qiagen Gel Purification kit with final elution in water.
2.2. Generation of transgenic Xenopus laevis tadpoles
All experimental procedures involving the use of frogs were carried out in accordance with the protocol approved by the University of Iowa Animal Care and Use Committee and compliant with the ARRIVE guidelines. Transgenic tadpoles were produced through the process of microinjections in which transgenes were introduced into decondensed sperm nuclei using the method of restriction enzyme mediated integration (REMI) (Kroll and Amaya, 1996). In REMI, sperm nuclei and egg extract were incubated with linearized plasmid and XhoI. The restriction enzyme enables minor cuts in the sperm genomic DNA, and linear plasmid gets incorporated into sperm genomic DNA during DNA repair process (Amaya and Kroll, 2010). The nuclei were then microinjected into unfertilized eggs and successful embryos were maintained at 18–22°C until they develop to tadpoles of ~stage 50 (~15 days post-injection). Transgenic tadpoles were identified by visual examination for EGFP fluorescence in tadpole eyes using an MZ16F Leica fluorescence microscope with a GFP filter.
2.3 Live cell imaging and cryo-sectioning
Transgenic tadpoles were initially anesthetized for 5 min in 0.02% Tricaine and transferred to Ringer buffer (10 mM HEPES (pH 7.5), 110 mM NaCl, 2 mM CaCl2, 2.5 mM KCl and 1 mM MgCl2). Retinas were extracted from the eyeball and minced in 60 μl of Ringer buffer on a glass slide using two 30-G needles. EGFP fluorescence in living cells was imaged immediately using an LSM 510 confocal microscope (Zeiss). Simultaneously, anesthetized tadpoles were fixed in 4% paraformaldehyde for 1hr. Later, tadpoles were incubated in 30% sucrose in PBS and 30% sucrose and OCT (1:1) solution, each for 1hr. Tadpole heads were dissected and embedded in OCT and frozen at −80°C until use. Cryo-sections were made using Leica Microm Cryostat HM505E and stored at −80°C until use.
2.4. Staining and confocal microscopy
Frozen sections were thawed to room temperature and washed in PBS twice for 5min. For staining rod OS, sections were incubated in 0.2% Triton in PBS for 30 min and stained with 2 μg/ml Alexa Fluor Wheat Germ Agglutinin 594 conjugates (Molecular Probes, Invitrogen) (WGA) dissolved in 0.2% Triton-PBS for 30min. WGA is commonly used to visualize rod OS (Luo et al., 2004, Karan et al., 2011). Sections were washed twice with 0.2% Triton-PBS and twice with PBS only. For nuclear counter-staining, sections were treated with 5μg/ml RNase A (Thermo Scientific) in PBS for 5 min followed by wash with PBS twice, 5min each. Later, sections were incubated with 1 μM TO-PRO3 (Invitrogen) solution in PBS. TO-PRO3 nuclear counter-staining combined with WGA staining of OS facilitates identification of rod inner segments (IS). Slides were washed briefly in PBS and mounted using Vectashield mounting medium (Vector Labs, Burlingame, CA). Sections were imaged using a LSM510 confocal microscope (Zeiss).
2.5. Immunohistochemistry
Frozen sections were thawed to room temperature and incubated in 0.2% Triton in PBS for 30 min followed by wash in PBS, for twice, 5 min each. Sections were incubated with anti-EE monoclonal antibody (Covance) (1:500 dilution) dissolved in 3%BSA in PBS for 1 hr with gentle shaking and were washed with PBS twice, 5min each. Later, sections were incubated with goat anti-mouse Alexa Fluor 568 secondary antibody (1:1000 dilution) for 1 hr with gentle shaking and washed with PBS twice, 5 min each. Subsequently, sections were stained with 1μM TO-PRO3 (Invitrogen) solution in PBS. Slides were washed briefly in PBS and mounted using Vectashield mounting medium (Vector Labs, Burlingame, CA).
2.6. Protein extraction and Immunoblotting
Eyeballs were excised from tadpoles at 2 weeks of age and stored at −80°C until use. Typically, around 100 eyeballs were homogenized with pestle in 1.5 ml tubes (Beckman Microfuge Polyallomer) using 200 μl of Buffer A (20 mM Tris-HCl (pH 7.5), 120 mM NaCl, 1 mM MgSO4, 1 mM 2-mercaptoethanol, and Complete-Mini EDTA-free protease inhibitor tablets (Roche)). The homogenate was centrifuged at 20,000xg and 4°C for 20 min. and supernatant was termed as the isotonic extract. The resulting pellet was resuspended in 200 μl of Buffer B (10 mM Tris-HCl (pH 7.5), 1 mM 2-mercaptoethanol and Complete-Mini EDTA-free protease inhibitor tablets (Roche)). The solution was centrifuged at 70,000xg and 4°C for 1hr. This supernatant was termed as the hypotonic extract. Samples equivalent to 10–12 eyeballs were subjected to SDS-PAGE in 4–12% pre-cast gels (Invitrogen) and electro-transferred onto nitrocellulose membrane using iBLOT dry transfer method (Invitrogen). Membranes were incubated with anti-GFP B-2 monoclonal antibody (Santa Cruz Biotech) (1:1000 dilution) or with anti-EE monoclonal antibody (Covance) (1:1000 dilution) at 4°C overnight. The antibody-antigen complexes were detected using anti-mouse antibodies conjugated to horseradish peroxidase (Santa Cruz Biotech) (1:10,000 dilution) and ECL Prime reagent (Amersham Pharmacia Biotech).
3. Results
3.1. PDE6C with ACHM mutations in the catalytic domain are properly transported to the OS
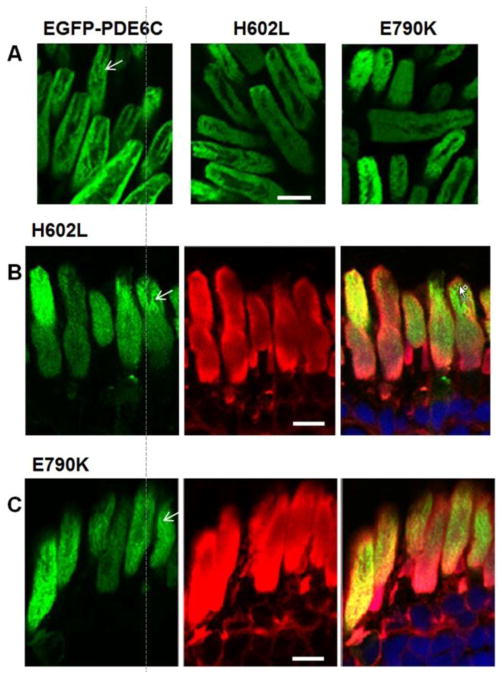
Seven missense mutations of PDE6C linked to ACHM fall into three categories based on their sequence localization (Fig. 1). Two mutations, H602L and E790K, are situated in the C-terminal catalytic domain. Three mutations are within the regulatory N-terminal GAF domains, R104W in GAFa and Y323N and P391L in GAFb. Two mutations, R29W in the N-terminus and M455V in the region linking GAFb to the catalytic domain, are located outside the conserved domains. First, we examined expression and trafficking of the catalytic domain mutants. Human EGFP-PDE6C localizes to the OS in rods of transgenic X. laevis in a characteristic striated pattern (Muradov et al., 2009). Live cell imaging and imaging of retina cryosections from the H602L and E790K transgenic tadpoles revealed that both PDE6C mutants were targeted to the OS (Fig. 2). Moreover, the pattens of H602L and E790K distribution in the OS were similar to that previously observed for EGFP-PDE6C.
Figure 1. Achromatopsia-linked missense mutations of PDE6C mapped on the transgenic construct for expression in rods of X. laevis.
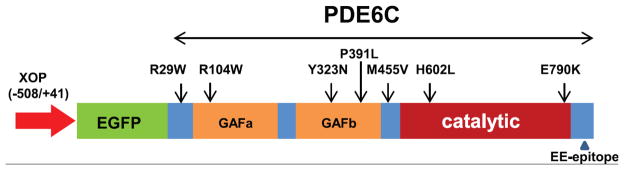
XOP – Xenopus opsin promoter.
Figure 2. The catalytic domain ACHM mutants of PDE6C are properly targeted to the OS.
(A) Live cell imaging of EGFP-PDE6C, H602L, and E790K. (B, C) Cryosection imaging of H602L and E790K indicate that the catalytic domain mutants of PDE6C are compartmentalized in the OS in the striated pattern (indicated by arrows). EGFP-fluorescence – green, WGA-staining – red, and the overlay combined with the channel for TO-PRO3 nuclear stain (blue). Scale bar - 10 μm.
Due to its isoprenylation, PDE6 is a peripheral membrane protein in isotonic solution, which can be solubilized by hypotonic buffers (Baehr et al., 1979). Similarly to native PDE6, EGFP-PDE6C in transgenic frog rods is extracted from the membrane with hypotonic buffer (Fig. 3). Sequential isotonic and hypotonic extractions of transgenic eyeballs combined with immunoblotting were performed to assess expression and the membrane binding properties of the H602L and E790K mutants. The EGFP-fused full-length H602L and E790K proteins (~130 kDa bands) were found only in the hypotonic extract fractions at comparable levels (Fig. 3). This analysis also revealed an additional EGFP-containing band of ~70 kDa present in the H602L and E790K extracts. This band was relatively minor for H602L and more prominent for E790K. Apparently, the 70 kDa band is a product of proteolysis, most likely at a single site. Thus, H602L and E790K exhibit a small and a moderate reduction in proteolytic stability, respectively.
Figure 3. Different proteolytic stability of the ACHM-linked mutants of PDE6C.
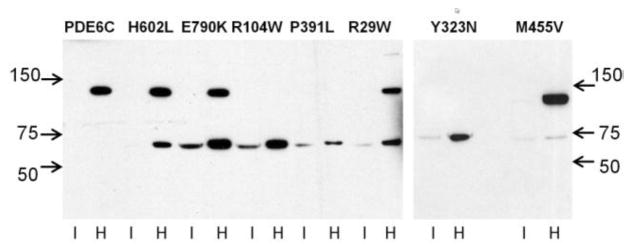
EGFP-PDE6C and mutant PDE6C proteins were extracted from transgenic eyeballs with isotonic (I) and hypotonic buffer (H) and analyzed by immunoblotting with anti-GFP antibody. The full-length EGFP-PDE6C and mutants correspond to the ~130 kDa band. The ~70 kDa band represents an N-terminal proteolytic fragment of PDE6C fused to EGFP.
3.2. ACM mutations in the GAF domains of PDE6C lead to a discrete proteolysis of the enzyme
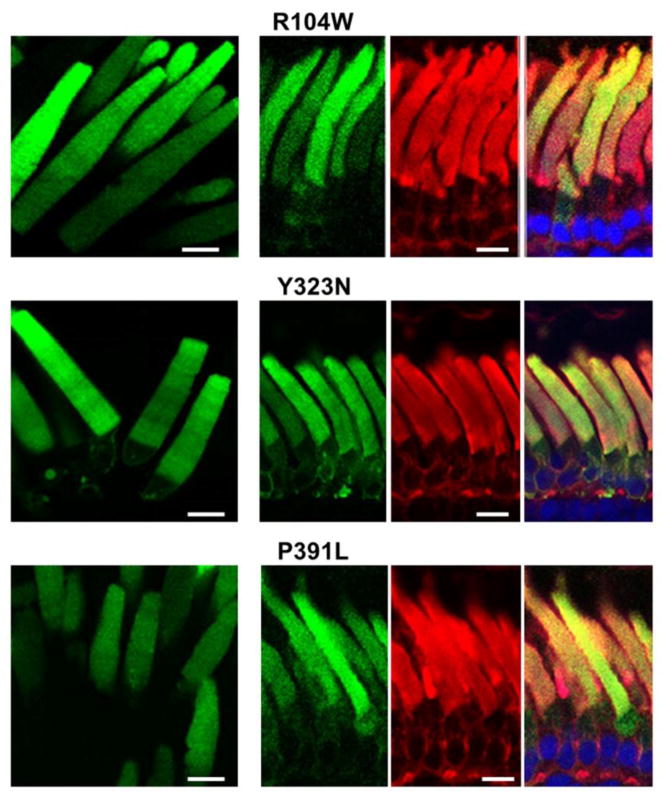
The PDE6C mutants of the regulatory GAF domains, R104W, Y323N, and P391L, showed analogous localization patterns in transgenic rods. For the three mutants, EGFP-fluorescence was observed mainly in the OS (Fig. 4). However, the distribution pattern of R104W, Y323N and P391L in the OS was diffused, which is different from the striated distributions of EGFP-PDE6C, H602L and E790K. Similarly to EGFP-PDE6C, expression of the mutants in transgenic X. laevis was mosaic (Muradov et al., 2009), i.e. expression levels varied between transgenic tadpoles and between different rods within a single retina. However, the diffused pattern of the GAF-domain mutants was the same in weakly or brightly fluorescent rods, suggesting that the pattern was not influenced by transgene expression levels. The membrane extraction and immunoblotting revealed only a ~70 kDa band containing EGFP band for R104W, Y323N, and P391L (Fig. 3). The GAF domain mutants were cleaved completely as the full-length 130 kDa proteins were not detected. Since complete cleavage was observed for the mutant with average expression comparable to that of EGFP-PDE6C (R104W) or significantly lower (P391L) (Fig. 3), the degree of proteolysis does not appear to be dependent on mutant expression levels. The regulatory N-terminal portion of PDE6C fused to EGFP appears to retain correct folding as it was trafficked to the OS.
Figure 4. Subcellular compartmentalization of the GAF domain ACHM mutants of PDE6C.
Live cell imaging (left) and cryosection imaging (right panels) of the GAF domain mutant EGFP-PDE6C proteins R104W, Y323N, and P391L. EGFP-fluorescence – green, WGA-staining – red, and the overlay combined with the channel for TO-PRO3 nuclear stain (blue). Scale bar - 10 μm.
3.3. Mutations outside conserved domains mislocalize PDE6C inside the OS
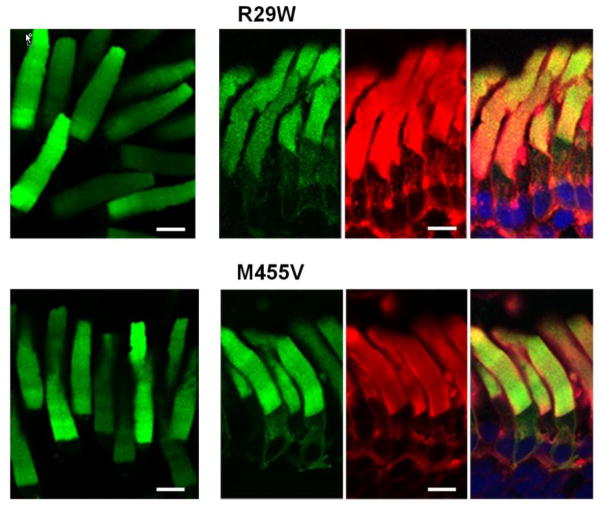
A multiple sequence alignment of rod and cone PDE6 enzymes from various species (not shown) indicates that among the two mutated residues outside the structural domains of PDE6C, M455 is more conserved. On the other hand, the R29W substitution may produce a larger impact compared to M455V as it replaces a positively charged residue with a bulky hydrophobic residue. Upon expression in transgenic X. laevis rods, both mutants localized primarily to the OS (Fig. 5). The distribution of R29W and M455V in the OS was diffused, similarly to the EGFP pattern of the GAF domain mutants (Figs. 4, 5). Yet, in contrast to the GAF domain mutants, M455V was resistant to proteolysis, whereas R29W showed partial proteolytic cleavage to the 70 kDa product (Fig. 3). Thus, the lack of striated pattern for R29W and M455V may reflect a specific mislocalization defect.
Figure 5. Diffused OS localization of the PDE6C proteins with mutations outside the conserved domains.
Live cell imaging (left) and cryosection imaging (right panels) of R29W and M455V. EGFP-fluorescence – green, WGA-staining – red, and the overlay combined with the channel for TO-PRO3 nuclear stain (blue). Scale bar - 10 μm.
3.4. Localization of the catalytic fragment(s) of PDE6C
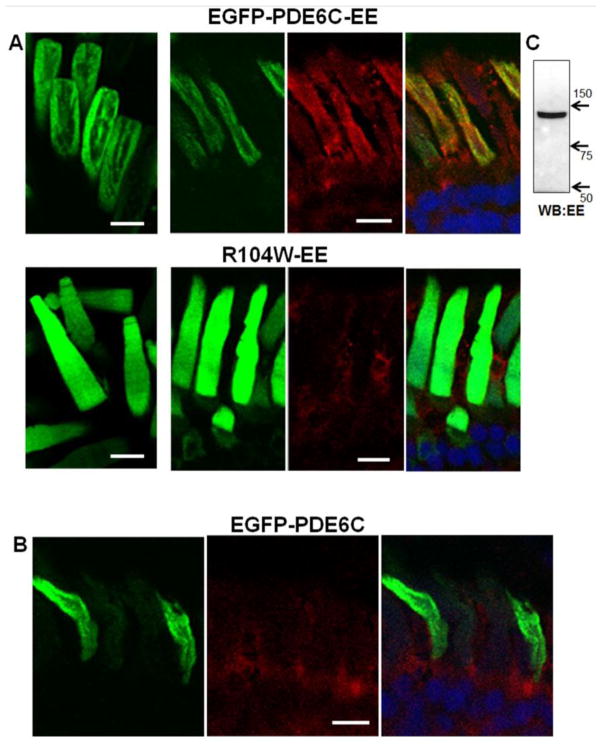
Proteolytic cleavage of mutant PDE6C, particularly the GAF domain mutants, resulted in a ~70 kDa fragment suggesting that it contains ~350–400 N-terminal aa of PDE6C fused to EGFP. Thus, the C-terminal proteolytic fragment(s) of PDE6C can be as large as 450 aa or more. To examine the proteolytic stability and trafficking of the C-terminal fragment, we introduced an epitope for monoclonal EE antibody at about 20 aa upstream of the PDE6C C-terminus (Fig. 1). The EE epitope did not interfere with the expression and correct OS targeting of EGFP-PDE6C-EE. The OS localization of EGFP-PDE6C-EE was confirmed by EGFP-fluorescence and EE-immunofluorescence (Fig. 6A). Control EE-staining of EGFP-PDE6C cryosections showed only weak nonspecific signal in the IS (Fig. 6B). The imaging of R104W-EE mutant revealed the same pattern of diffused EGFP-fluorescence in the OS as it was observed for this mutant lacking the EE-epitope (Fig. 6A). However, no EE-immunofluorescence was detected in the OS, suggesting that the catalytic fragment(s) is absent from this compartment (Fig. 6A). Furthermore, while immunoblotting of EGFP-PDE6C-EE extract with anti-EE-antibodies showed the expected 130 kDA band (Fig. 6C), no EE-specific bands were detected in R104W-EE extract (not shown).
Figure 6. Imaging of PDE6C and R104W with the C-terminal EE-epitope.
A. Live cell imaging (left) and cryosection immunohistochemistry (right panels) of EGFP-PDE6C-EE and R104W-EE. B. Cryosection immunohistochemistry of EGFP-PDE6C. EGFP-fluorescence – green, EE-immunofluorescence – red, and the overlay combined with the channel for TO-PRO3 nuclear stain (blue). Scale bar - 10 μm. C. Immunoblot analysis of EGFP-PDE6C-EE hypotonic extract (sample equivalent to 40 eyeballs) with anti-EE monoclonal antibody.
4. Discussion
The mechanisms of PDE6 mutations leading to severe retinal diseases are poorly understood, particularly since a system for expression of functional PDE6 has been lacking. This impediment was principally overcome with the demonstration that transgenic X. laevis is an excellent tool for expression and studies of PDE6 in living rods (Muradov et al., 2009, Muradov et al., 2010). Additionally, biochemical analyses of recombinant PDE6 in vitro are feasible following selective immunoprecipitation of the enzyme from transgenic retina extracts (Muradov et al., 2010). Biochemical studies on large sets of PDE6 mutants are not practical due to the need to produce large numbers of transgenic tadpoles. In contrast, imaging combined with membrane extraction and immunoblotting can be advantageous in characterization of multiple PDE6 mutants. Recently, we used this methodology to identify a novel GAFa domain-dependent OS localization signal of PDE6 (Cheguru et al., 2014). In this study, this approach was applied to gain insights into the mechanisms of missense mutations in PDE6C causing ACHM in human patients.
Seven ACHM-linked mutants of PDE6C expressed in rods of transgenic X. laevis revealed differing patterns of proteolytic stability and compartmentalization. Similarly to EGFP-PDE6C, two catalytic domain mutants, H602L and E790K, were compartmentalized in the rod OS in the striated pattern. Also, the full-length H602L and E790K associated with the membrane in the manner analogous to the membrane binding of EGFP-PDE6C. These results suggest that H602L and E790K are for the most part correctly folded and able to properly traffic in rods. Some level of proteolysis of H602L and E790K may be indicative of modest destabilization of the proteins. The key mechanism of H602L in ACHM appears to be its catalytic deficiency. Two highly conserved metal-binding motifs in PDEs, HX3HXnE, are critical for coordination Zn2+ and Mg2+ and indispensable in the hydrolysis of cyclic nucleotides (Conti and Beavo, 2007). H602 corresponds to the first His in the first metal binding site. Mutation of the H602 counterpart in PDE5 leads to a profound reduction in PDE5 activity (Turko et al., 1998). The structures of PDE5 indicate that the His residue is not directly involved in coordination of the metals, but rather it influences the topology of the metal-binding site (Sung et al., 2003). This may explain the absence of significant protein destabilization in the H602L mutant. The normal trafficking of H602L suggests that catalytic activity of PDE6 and its transport to the OS are not linked. The mechanism of the E790K mutation is less clear, but it may involve a combination of several factors. Although the E790K mutation is not likely to affect the enzyme catalytic site directly, the mutant appears to be somewhat less stable than the H602L mutant. In addition, the E790K transcript processing is impaired (Grau et al., 2011). Reduced expression of E790K due to abnormal transcript processing coupled with lower protein stability may decrease basal PDE6 activity below the threshold level triggering ACHM.
Mutations in the GAF domains of PDE6, R104W, Y323N, P391L, led to impaired protein stability as judged by the proteolytic cleavage of the mutants. Surprisingly, the proteolysis of the PDE6C mutants produced a single EGFP-containing product of ~70 kDa. This proteolytic pattern suggests a structural pertubation that initially exposes a single proteolytically-sensitive site. Based on the size of the proteolytic fragment, the cleavage occurs within the C-terminal portion of the GAFb domain (aa ~350–400). The homology model of the GAFa-GAFb dimer of PDE6C based on the atomic structure of PDE5 shows an exposed and largely unstructured loop corresponding to ~aa 356–386 (Fig. 7). This loop appears to be a strong candidate for the proteolytic cleavage site. From the homology model, the P391L substitution may directly affect the conformation of the loop through the P391 interactions with the backbone at aa 356GFI358 (Fig. 7). The proteolytic cleavage in the GAFb domain caused by the GAFa mutation R104W possibly results from the disruption of the interdomain contacts. In the model, R104 comes to close proximity with the GAFb domain, while the neighboring N105 is within a hydrogen-bonding distance with the GAFb aa 415DEH417 (Fig. 7). Residue corresponding to R104 is highly conserved among cone and rod PDE6. The R104W mutation was homozygous in ACHM patients (Chang et al., 2009). Interestingly, mutations of this residue in PDE6A, R102H and R102S, cause RP in humans (Dryja et al., 1999). The mechanism of RP due to these PDE6A mutations is expected to be similar to the mechanism of R104W in ACHM. Subcellular localization and identity of the protease that is cleaving mutant PDE6C proteins is unknown. We think that the cleavage is likely to occur in the IS. The subsequent trafficking of the large N-terminal fragment of PDE6C to the OS is in agreement with the reported OS targeting of EGFP-GAFa fusion proteins (Cheguru et al., 2014). The C-terminal catalytic fragment of PDE6C does not traffic to the OS, and apparently, undergoes proteolytic degradation in the IS.
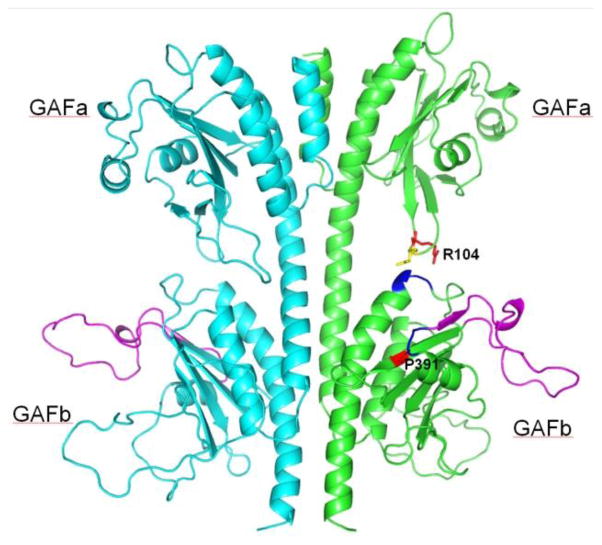
Figure 7. Model of the GAFa-GAFb domains of PDE6C.
The homology model of the PDE6C GAFa-GAFb domain dimer was generated using SWISS-MODEL (Arnold et al., 2006) and the structure of the PDE5 GAF-domain dimer (PDB ID: 3LFV) (Wang H et al., 2010). The probable proteolytic cleavage site in EGFP-PDE6C leading to ~70 kDa N-terminal fragment (loop aa 359–386) is shown in magenta. R104 and P391 are shown in red, and N105 is yellow. The backbone of aa 356GFI358 and 415DEH417 making contacts with P391 and N105, respectively, is shown in blue.
Lastly, mutations R29W and M455V residing outside the conserved domains of PDE6C displayed a distinct pattern of protein compartmentalization. Similarly to H602L and E790K, the full-length R29W and M455V mutant proteins were transported to the OS. However, unlike H602L and E790K, the OS distribution of R29W and M455V was diffused rather than striated, suggesting that the mutants failed to localize to the disc rim regions as EGFP-PDE6C. It remains to be investigated how such mislocalization affects PDE6C function and causes ACHM. Overall, our results suggest a spectrum of mechanisms of missense PDE6C mutations in ACHM that involves catalytic defects, protein mislocalization, or the initial cleavage in the GAFb domain followed by proteolytic degradation of the large C-terminal fragment in the IS.
Highlights.
Achromatopsia-linked mutants of cone PDE6 were expressed in rods of transgenic frogs.
PDE6 mutants were examined for their stability and compartmentalization.
A spectrum of mechanisms of missense PDE6C mutations in achromatopsia is proposed.
Acknowledgments
We would like to thank Dr. Sheila Baker (University of Iowa) for helpful comments on the manuscript, Dr. Hakim Muradov for construction of several PDE6C mutants and Kimberly Boyd for technical assistance. This work was supported by National Institutes of Health Grant EY-10843 to N.O.A.
Abbreviations
- PDE6
photoreceptor phoshphodiesterase-6
- PDE5
cGMP-binding, cGMP-specific phoshphodiesterase-5
- ACHM
autosomal recessive achromatopsia
- GAF
domains, termed for their presence in cGMP-regulated PDEs, adenylyl cyclases, and the E. coli protein Fh1A
- OS
outer segment
- IS
inner segment
- WGA
Wheat Germ Agglutinin
- DTT
dithiothreitol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amaya E, Kroll K. Production of transgenic Xenopus laevis by restriction enzyme mediated integration and nuclear transplantation. Journal of visualized experiments: JoVE. 2010 doi: 10.3791/2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anant JS, Ong OC, Xie HY, Clarke S, O’Brien PJ, Fung BK. In vivo differential prenylation of retinal cyclic GMP phosphodiesterase catalytic subunits. J Biol Chem. 1992;267:687–690. [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Arshavsky VY, Burns ME. Photoreceptor signaling: supporting vision across a wide range of light intensities. J Biol Chem. 2012;287:1620–1626. doi: 10.1074/jbc.R111.305243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehr W, Devlin MJ, Applebury ML. Isolation and characterization of cGMP phosphodiesterase from bovine rod outer segments. J Biol Chem. 1979;254:11669–11677. [PubMed] [Google Scholar]
- Bowes C, Li T, Danciger M, Baxter LC, Applebury ML, Farber DB. Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature. 1990;347:677–680. doi: 10.1038/347677a0. [DOI] [PubMed] [Google Scholar]
- Catty P, Pfister C, Bruckert F, Deterre P. The cGMP phosphodiesterase-transducin complex of retinal rods. Membrane binding and subunits interactions. J Biol Chem. 1992;267:19489–19493. [PubMed] [Google Scholar]
- Chang B, Grau T, Dangel S, Hurd R, Jurklies B, Sener EC, Andreasson S, Dollfus H, Baumann B, Bolz S, Artemyev N, Kohl S, Heckenlively J, Wissinger B. A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc Natl Acad Sci U S A. 2009;106:19581–19586. doi: 10.1073/pnas.0907720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheguru P, Zhang Z, Artemyev NO. The GAFa domain of phosphodiesterase-6 contains a rod outer segment localization signal. J Neurochem. 2014;129:256–263. doi: 10.1111/jnc.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- Dryja TP, Rucinski DE, Chen SH, Berson EL. Frequency of mutations in the gene encoding the alpha subunit of rod cGMP-phosphodiesterase in autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1999;40:1859–1865. [PubMed] [Google Scholar]
- Farber DB, Lolley RN. Cyclic guanosine monophosphate: elevation in degenerating photoreceptor cells of the C3H mouse retina. Science. 1974;186:449–451. doi: 10.1126/science.186.4162.449. [DOI] [PubMed] [Google Scholar]
- Fu Y, Yau KW. Phototransduction in mouse rods and cones. Pflugers Arch. 2007;454:805–819. doi: 10.1007/s00424-006-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau T, Artemyev NO, Rosenberg T, Dollfus H, Haugen OH, Cumhur Sener E, Jurklies B, Andreasson S, Kernstock C, Larsen M, Zrenner E, Wissinger B, Kohl S. Decreased catalytic activity and altered activation properties of PDE6C mutants associated with autosomal recessive achromatopsia. Hum Mol Genet. 2011;20:719–730. doi: 10.1093/hmg/ddq517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karan S, Tam BM, Moritz OL, Baehr W. Targeting of mouse guanylate cyclase 1 (Gucy2e) to Xenopus laevis rod outer segments. Vision Res. 2011;51:2304–2311. doi: 10.1016/j.visres.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karan S, Zhang H, Li S, Frederick JM, Baehr W. A model for transport of membrane-associated phototransduction polypeptides in rod and cone photoreceptor inner segments. Vision Res. 2008;48:442–452. doi: 10.1016/j.visres.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Liu X, Bulgakov OV, Wen XH, Woodruff ML, Pawlyk B, Yang J, Fain GL, Sandberg MA, Makino CL, Li T. AIPL1, the protein that is defective in Leber congenital amaurosis, is essential for the biosynthesis of retinal rod cGMP phosphodiesterase. Proc Natl Acad Sci U S A. 2004;101:13903–13908. doi: 10.1073/pnas.0405160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Marsh-Armstrong N, Rattner A, Nathans J. An outer segment localization signal at the C terminus of the photoreceptor-specific retinol dehydrogenase. J Neurosci. 2004;24:2623–2632. doi: 10.1523/JNEUROSCI.5302-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP. Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci U S A. 1995;92:3249–3253. doi: 10.1073/pnas.92.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muradov H, Boyd KK, Artemyev NO. Rod phosphodiesterase-6 PDE6A and PDE6B subunits are enzymatically equivalent. J Biol Chem. 2010;285:39828–39834. doi: 10.1074/jbc.M110.170068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muradov H, Boyd KK, Haeri M, Kerov V, Knox BE, Artemyev NO. Characterization of human cone phosphodiesterase-6 ectopically expressed in Xenopus laevis rods. J Biol Chem. 2009;284:32662–32669. doi: 10.1074/jbc.M109.049916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natochin M, Granovsky AE, Artemyev NO. Identification of effector residues on photoreceptor G protein, transducin. J Biol Chem. 1998;273:21808–21815. doi: 10.1074/jbc.273.34.21808. [DOI] [PubMed] [Google Scholar]
- Pittler SJ, Baehr W. Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase beta-subunit gene of the rd mouse. Proc Natl Acad Sci U S A. 1991;88:8322–8326. doi: 10.1073/pnas.88.19.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy V, Niemi GA, Reh TA, Hurley JB. Leber congenital amaurosis linked to AIPL1: a mouse model reveals destabilization of cGMP phosphodiesterase. Proc Natl Acad Sci U S A. 2004;101:13897–13902. doi: 10.1073/pnas.0404197101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung BJ, Hwang KY, Jeon YH, Lee JI, Heo YS, Kim JH, Moon J, Yoon JM, Hyun YL, Kim E, Eum SJ, Park SY, Lee JO, Lee TG, Ro S, Cho JM. Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules. Nature. 2003;425:98–102. doi: 10.1038/nature01914. [DOI] [PubMed] [Google Scholar]
- Thiadens AA, den Hollander AI, Roosing S, Nabuurs SB, Zekveld-Vroon RC, Collin RW, De Baere E, Koenekoop RK, van Schooneveld MJ, Strom TM, van Lith-Verhoeven JJ, Lotery AJ, van Moll-Ramirez N, Leroy BP, van den Born LI, Hoyng CB, Cremers FP, Klaver CC. Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am J Hum Genet. 2009;85:240–247. doi: 10.1016/j.ajhg.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turko IV, Francis SH, Corbin JD. Potential roles of conserved amino acids in the catalytic domain of the cGMP-binding cGMP-specific phosphodiesterase. J Biol Chem. 1998;273:6460–6466. doi: 10.1074/jbc.273.11.6460. [DOI] [PubMed] [Google Scholar]
- Veske A, Orth U, Ruther K, Zrenner E, Rosenberg T, Baehr W, Gal A. Mutations in the gene for the β-subunit of rod photoreceptor cGMP-specific phosphodiesterase (PDEB) in patients with retinal dystrophies and dysfunctions. In: Anderson RE, et al., editors. Degenerative Diseases of the Retina. Plenum, New York: Springer US; 1995. pp. 313–322. [Google Scholar]
- Wang H, Robinson HHK. Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain. J Biol Chem. 2010;285:38149. doi: 10.1074/jbc.M110.141614. [DOI] [PMC free article] [PubMed] [Google Scholar]