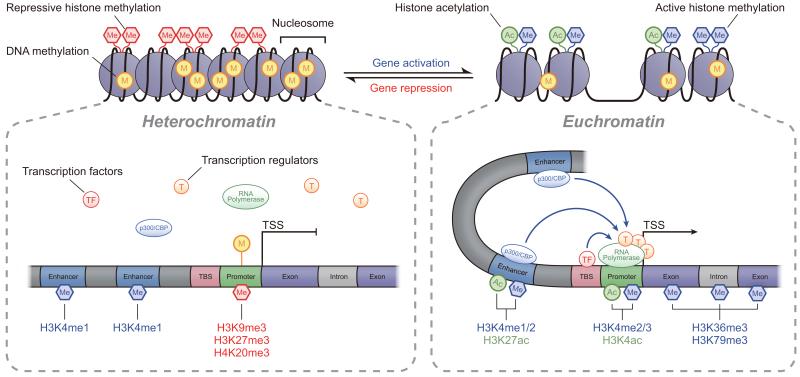
Fig. 2.
Chromatin structure and transcription regulation. Chromatin is made up of nucleosome subunits, each of which consists of an octamer of histones wrapped by chromosomal DNA. Lysine residues in the amino-terminal tails of nucleosomal histones are susceptible to several PTMs, including KAc and Kme, which regulate chromatin accessibility to transcription factors (TFs) and RNA polymerase II (labelled in the figure as RNA polymerase). Heterochromatin is marked by repressive histone PTMs, and increased DNAme at promoter CpG islands, leading to chromatin condensation and transcription repression. In contrast, euchromatin is enriched with permissive histone PTMs (H3/H4Kac and H3K4me1-3), and reduced promoter DNAme, leading to open chromatin and increased transcription. In addition, enhancers, which regulate promoters up to several thousand base pairs away, are marked by H3K4me1. Active enhancers are enriched with H3K27ac and the co-activator histone acetyl transferase p300. Actively transcribed gene bodies are enriched with H3K36me3/H3K79me3, whereas H3K27me3 is associated with reduced transcription. In addition, lncRNAs and miRNAs can also regulate gene expression by various post-transcriptional and epigenetic mechanisms (not shown here). TBS, TF binding site; TSS, transcription start site