Abstract
Higher sensitivity of NMR spectrometers and novel isotopic labeling schemes have ushered the development of rapid data acquisition methodologies, improving the time resolution with which NMR data can be acquired. For nucleic acids, longitudinal relaxation optimization in conjunction with Ernst angle excitation (SOFAST-HMQC) for imino protons, in addition to rendering rapid pulsing, has been demonstrated to yield significant improvements in sensitivity per unit time. Extending such methodology to other spins offers a viable prospect to measure additional chemical shifts, thereby broadening their utilization for various applications. Here, we introduce the 2D [13C,1H] aromatic SOFAST-HMQC that results in overall sensitivity gain of 1.4–1.7 fold relative to the conventional HMQC and can also be extended to yield long-range heteronuclear chemical shifts such as the adenine imino nitrogens N1, N3, N7 and N9. The applications of these experiments range from monitoring real-time biochemical processes, drug/ligand screening, and to collecting data at very low sample concentration and/or in cases where isotopic enrichment cannot be achieved.
Keywords: Longitudinal relaxation optimization, Ernst angle, SOFAST-HMQC, nucleic acids, fast NMR
The inherent insensitivity of NMR spectroscopy has been significantly alleviated with the widespread adoption of cryogenic probes (Kovacs et al. 2005) along with novel and economical isotope labeling schemes. This resulted in acquiring data in the sampling-limited regime (Szyperski et al. 2002) where the time required for sampling the indirect evolution points, rather than signal averaging, dictates the total measurement time and thus motivated development of methodology aimed at fast data acquisition (Atreya and Szyperski 2005; Felli and Brutscher 2009; Mishkovsky and Frydman 2009; Rennella and Brutscher 2013). NMR data can be rapidly acquired by reducing the time required to acquire indirect evolution points, such as in projection NMR spectroscopy (Szyperski et al. 1993; Kim and Szyperski 2003; Kupče and Freeman 2004; Eghbalnia et al. 2005; Hiller et al. 2005; Atreya et al. 2012; Krähenbühl et al. 2012), sparse sampling with non-FT based data processing (Brüschweiler and Zhang 2004; Rovnyak et al. 2004; Maciejewski et al. 2011; Mobli et al. 2012) and Hadamard NMR spectroscopy (Kupče and Freeman 2003). Alternatively, it can be achieved by reducing/eliminating the inter-scan delay between transients, as in spatially selective approaches (Frydman et al. 2002; Parish and Szyperski 2008; Giraud et al. 2010; Vega-Vazquez et al. 2010; Sathyamoorthy et al. 2014) and/or with Longitudinal relaxation (L-)optimization (Pervushin et al. 2002; Deschamps and Campbell 2006).
L-optimization offers an attractive rapid pulsing approach and has been extensively applied in protein NMR studies. It relies on selective excitation of the spins of interest, with the unperturbed spins acting as a “relaxation sink” via dipolar interactions or chemical exchange with solvent protons. This results in an effective reduction of T1 relaxation times of the excited spins, thereby permitting the use of smaller inter-scan delays. Combining L-optimization of protons with Ernst optimum flip angle excitation (Ernst et al. 1987) for HMQC (Ross et al. 1997) resulted in the band-Selective Optimized Flip Angle Short Transient (SOFAST-) HMQC (Schanda and Brutscher 2005) experiments that yield significant improvements in sensitivity.
Despite lower proton density relative to proteins (Farjon et al. 2009), SOFAST-HMQC type experiments have successfully been applied to nucleic acids by targeting the isolated and downfield shifted imino protons yielding 2-fold sensitivity boosts (Farjon et al. 2009). This includes L-optimized Transverse Relaxation Optimized SpectrscopY (TROSY) for studying large nucleic acids and HNN-COSY experiment that help in establishing the H-bonding partners in nucleic acids. In addition, 2D ultraSOFAST NMR, which combines SOFAST-HMQC with spatially selective single scan approach (Tal and Frydman 2010), made it possible to collect 2D [15N, 1H] imino HMQC spectra every few seconds and thereby monitor the real-time folding of the 71 nucleotide long add adenine-sensing riboswitch aptamer domain (Lee et al. 2010).
Imino 1H/15N chemical shifts, although abundant in information regarding base-pairing and secondary structure, are limited to only two out of the four bases and available only in cases where the imino proton is hydrogen-bonded and protected from solvent exchange such as in G-C/U/T and A-U/T base-pairs. Imino chemical shift data is often unavailable for non-canonical motifs, such as bulges, internal and apical loops, and many non-canonical base-pairs. These motifs play essential architectural roles defining global RNA 3D structure and often form the key sites involved in binding to proteins and ligands (Gallego and Varani 2001; Schwalbe et al. 2007; Dethoff et al. 2012; Reining et al. 2013). The non-exchangeable aromatic protons (H2, H6 and H8) and their directly bonded carbons (C2, C6, and C8) make it possible to more broadly access nucleobase chemical shift information in canonical and non-canonical residues. In addition, the nucleobase carbon (C2, C5, C6, and C8) and proton (H2, H5, H6, and H8) chemical shifts are increasingly being used in defining the 3D structures of RNA and DNA (Wijmenga et al. 1997; Cromsigt et al. 2001; Xu and Case 2001; Barton et al. 2013; Frank et al. 2013; Frank et al. 2013; Sahakyan and Vendruscolo 2013; Werf et al. 2013; Sripakdeevong et al. 2014). Having the ability to acquire these chemical shift data rapidly would be important for applications ranging from nucleic acid targeted ligand screening to time-resolved NMR studies of biochemical processes such as catalysis (Buck et al. 2009) and folding (Wenter et al. 2005; Buck et al. 2007; Manoharan et al. 2009; Lee et al. 2010; Lieblein et al. 2012; Li et al. 2014). In addition, L-optimization for aromatic protons has been successfully implemented to measure residual dipolar couplings for nucleic acids (Ying et al. 2011). This study demonstrated the apparent reduction of aromatic proton T1, owing to the fact that magnetization from water protons is transferred rapidly to the non-exchangeable aromatic protons via the exchangeable protons (amino, imino and hydroxyl), resulting in shorter inter-scan delays. This study also showed the advantage of using H2O against D2O as solvent due to favorable viscosity properties.
With the foundation for L-optimization of aromatic proton laid earlier (Ying et al. 2011), in this study we present the implementation of 2D [13C, 1H] aromatic SOFAST-HMQC and additionally, as a proof of concept, we demonstrate that the SOFAST-HMQC methodology can be extended to acquire other nucleobase 13C/15N chemical shifts. Application of these experiments are presented for a (a) 29 nucleotide 100% uniformly 13C/15N labeled transactivation response element RNA (TAR) from HIV type-1 (5′-GGCAGAUCUGAGCCUGGGAGCUCUCUGCC-3′), (b) 76 nucleotide in vitro transcribed 100% uniformly 13C/15N labeled phenylalanine transfer RNA (tRNAPhe) from organism Saccharomyces cerevisiae (primary sequence in supplementary information), (c) 12 base-pair natural isotopic abundance palindromic Dickerson dodecamer duplex DNA (DD-dsDNA, 5′-CGCGAATTCGCG -3′) and (d) 12 base-pair 100% uniformly 13C/15N labeled G, T DD-dsDNA engineered with a G-T mispair (DD-GT-dsDNA, 5′-CGCGAATTTGCG-3′, G-T mispair is underlined). See supplementary information for more details. All experiments are implemented for samples in 90/10% H2O/D2O to take advantage of the exchangeable protons that act as T1 sinks for the aromatic spins (Ying et al. 2011).
The pulse sequence for the 2D [13C, 1H] aromatic SOFAST-HMQC is shown in Fig 1. The aromatic 1H selective excitation and refocusing is achieved using PC9 (Kupče and Freeman 1994) and REBURP (Geen and Freeman 1991) pulses, respectively, as described previously (Schanda et al. 2005). The selective pulses are centered at 8.0 ppm encompassing a bandwidth of ±1.5 ppm, with the imino, hydroxyl, amino and aliphatic 1H spins acting as T1 relaxation sinks. For uniformly 13C/15N labeled samples homonuclear 13C-13C decoupling during t1-evolution is achieved by employing a band-selective WURST-2 (Kupče and Freeman 1996; Dayie 2005) pulse (ω1 ~1350 Hz). The Ernst flip angle for excitation of aromatic protons (120°) was obtained by optimizing for maximum signal intensity at short inter-scan delays (Fig S1) motivated towards rapid pulsing as described previously (Schanda et al. 2005). All experiments were performed at 25 °C for TAR, DD-dsDNA and DD-GT-dsDNA and at 37 °C for tRNAPhe using a Varian INOVA 800 MHz spectrometer equipped with a cryogenic 1H[13C,15N] probe (Table S1). NMR data were processed using the software NMRPipe (Delaglio et al. 1995) and analyzed using the program SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco).
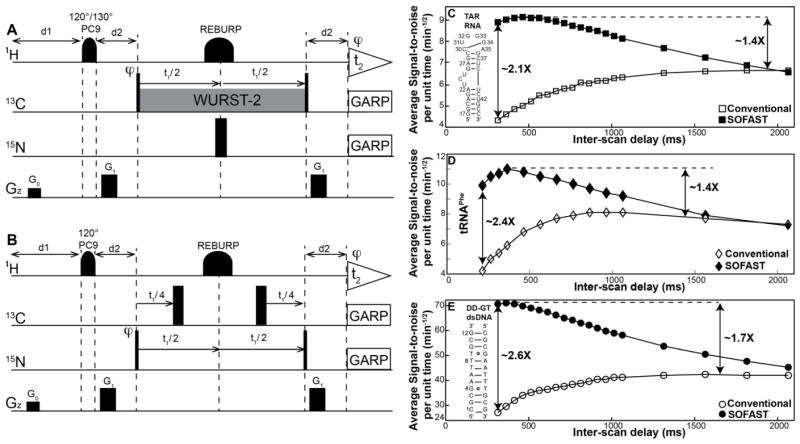
Fig 1.
(A) Pulse sequence for 2D aromatic [13C, 1H] SOFAST-HMQC for nucleic acids. (B) Pulse sequence for [15N, 1H] long-range SOFAST-HMQC experiment that correlates the two-bond N-H in purine bases of A and G. (A,B) Rectangular 90° and 180° pulses are indicated by thin and thick vertical bars, respectively, and phases are indicated above the pulses. The phase of all pulses is x unless indicated otherwise. PC9 and REBURP pulses are employed for selective excitation and refocusing of aromatic/imino protons. 120°/130° PC9 excitation pulses centered (with bandwidth) at 8.0 (±1.5 ppm) / 12.0–13.1 ppm (±2.0–3.0 ppm) are employed for aromatic/imino protons. The coherence transfer delay d2 is set to 2.5 ms corresponding to 1/(2JCH) for aromatic HMQC, while for the long-range 2D [15N, 1H] and [13C, 1H] SOFAST-HMQC experiments is optimized to 19.5 and 31.3 ms, respectively, for maximum signal intensity accounting for relaxation loses. Watergate sequence (Piotto et al. 1992) is employed with the selective 180° refocusing pulse for water suppression. Band selective WURST-2 pulses (ω1 ~1350 kHz) are used for homonuclear 13C-13C decoupling centered at 98 ppm for RNA (at 100 and 115 ppm for DNA) and at 15 and 150 ppm for the long-range [13C, 1H] experiment. Heteronuclear 13C-15N decoupling is accomplished by applying a 180° non-selective refocusing pulse is applied on the 15N channel with the carrier place at 200 and 235 ppm for aromatic and long range [13C, 1H] HMQC, respectively. Delayed acquisition (Cavanagh et al. 2006) is employed for t1-sampling, aiding in distinguishing folded peaks as they appear with opposite phase to that of regular peaks. A globally optimized alternating phase rectangular pulse (GARP) (Shaka et al. 1985) is employed to decouple 13C/15N (ω1 = 2.4/1 kHz) during signal acquisition. Pulsed z-field gradients (PFGs) are of rectangular shape, and their duration and strengths are: G0 (100 μs, 21 G/cm); G1 (600 μs, 24 G/cm). Phase cycling: φ = x, −x was employed for axial peak suppression (Cavanagh et al. 2006). Quadrature detection in t1(13C/15N) is accomplished by altering the phase φ according to States-TPPI (Cavanagh et al. 2006). Average signal-to-noise per unit time (SNt) as a function of inter-scan delay for (C) 1.0 mM uniformly 13C/15N labeled TAR RNA (D) 0.5 mM uniformly 13C/15N labeled tRNAPhe and (E) 3.0 mM uniformly 13C/15N labeled DD-GT-dsDNA from aromatic conventional (open objects) and SOFAST- (solid objects) HMQC experiments.
To assess the sensitivity gain offered by the aromatic SOFAST-HMQC experiment, the average signal-to-noise ratio per unit time (SNt) was measured as a function of the inter-scan delay (sum of recycle delay d1 and acquisition time t2) and compared with conventional HMQC experiment (Figs 1C, 1D and 1E). SNt is defined as the ratio of signal-to-noise ratio (SN) to the square root of measurement time, with the noise measured from representative 2D regions of the spectrum. The maximum SNt occurs at inter-scan delay of 300–450 ms for aromatic protons H2/H6/H8 achieving an average 1.4–1.7 fold sensitivity gain for nucleic acids in comparison to conventional HMQC. A similar gain observed for 26 kDa tRNAPhe indicates that the L-optimization strategy is an effective approach for rapid data acquisition for larger molecular weight systems as well. The H2 spins in particular showed a slightly lower average gain of 1.25–1.4 times as they are isolated from the remaining spins for effective longitudinal relaxation. While imino 1H spins have been reported to show the maximum SNt at an inter-scan delay of 200–300 ms (Farjon et al. 2009), the observed difference for the aromatic protons could arise as relatively less proximal T1 sinks are available and also that the H1′ aliphatic and H5 aromatic protons, contributors towards the longitudinal relaxation of aromatic protons, also get partially excited resulting in the need for a moderately longer inter-scan delay. It is important to note that the implementation of SOFAST-HMQC for H1′ sugar protons resulted in a much lower sensitivity boost (5–10%) due to the required water suppression which also excites the remaining aliphatic protons (H3′, H4′ and H5′/H5″) affecting the available T1 sinks, and therefore is not discussed further here.
In order to illustrate the application of the aromatic SOFAST-HMQC experiment in rapid data acquisition, Figs 2A, 2C and 2D show the high resolution 2D spectra of 1 mM uniformly 13C/15N labeled TAR (SN ~ 20), 0.5 mM uniformly 13C/15N labeled tRNAPhe (SN ~9) and ~3.5 mM palindromic DD-dsDNA (SN ~ 7) recorded at natural abundance acquired with a total measurement time of 19 seconds, 32 seconds and 2.5 minutes, respectively (Table S1). Comparison of rapidly acquired SOFAST-HMQC data with conventional HMQC spectra reveals that no artifacts are generated in the implemented pulse sequence (Figs S2 A, B). With the recent developments in sparse sampling methodologies (Hyberts et al. 2012; Mayzel et al. 2014), it should be possible to further reduce the total measurement time required to acquire the 2D data (Figs S2 A, C–E). The aromatic SOFAST-HMQC experiment therefore paves the way for carrying out time-resolved NMR studies of nucleic acids with a much larger number of chemical shift probes on all four nucleotide bases.
Fig 2.
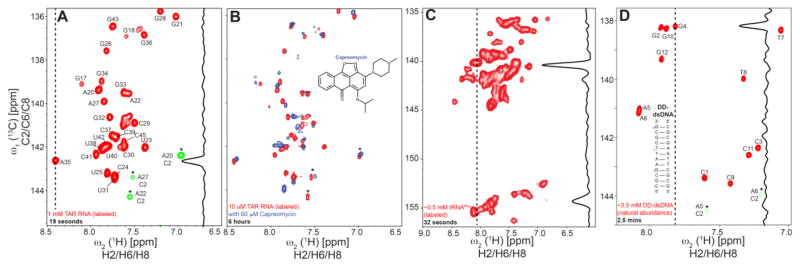
Aromatic 2D SOFAST-HMQC spectra acquired for 1 mM (A) and 10 μM (B) samples of uniformly 13C/15N labeled TAR, with a total measurement time of 19 seconds and 6 hours, respectively. The 1D trace across 1(13C) for A35 in (A) reflects the high intrinsic sensitivity of the experiment. The relatively lower sensitivity of the adenine C2-H2 peaks, folded over in the chosen spectral window (peaks denoted with *), indicate the longer T1 experienced by H2 spins as against H6/H8. In (B) the binding of Capreomycin, a small molecule computationally predicted to bind TAR RNA, to 10 μM uniformly 13C/15N labeled TAR sample is shown (in blue) illustrating the applicability of aromatic SOFAST-HMQC towards small molecule screening and also in sensitivity-limited cases. Folded peaks are denoted by (*) and colored same to maintain simplicity. (C) and (D) 2D aromatic SOFAST-HMQC spectra acquired within a measurement time of 32 seconds and 2.5 minutes for 0.5 mM uniformly 13C/15N labeled tRNAPhe and 3.5 mM unlabeled palindromic DD-dsDNA, respectively.
The sensitivity gain offered by L-optimization also greatly enables NMR-based drug/ligand screening against nucleic acid targets. Here, the ability to rapidly measure 2D [13C, 1H] aromatic HMQC in addition to imino 2D [15N, 1H] HMQC provides a more robust approach for monitoring ligand binding, as well as for carrying out detailed chemical shift mapping to characterize the binding site and for evaluating binding modes predicted using computational docking (Stelzer et al. 2011). This implementation also makes it possible to acquire 2D data at much lower RNA concentrations, which is often necessary due to poor solubility of tested ligands in aqueous solvents. Screening ligand-RNA binding by NMR is attractive not only due to the rich information that it can provide regarding binding affinity and binding mode but also because small molecules often absorb light making it difficult to assess binding using optical methods such as fluorescence. To demonstrate the applicability of the aromatic SOFAST-HMQC towards binding studies, Fig 2B shows the high-resolution 2D data of a 10 μM sample of uniformly 13C/15N labeled TAR with and without capreomycin, a small molecule hit obtained from a recent computational screening of TAR, acquired within a total measurement time of 6 hours per 2D spectra (Table S1) which would otherwise require ~18 hours with conventional HMQC.
The sensitivity gain achieved with the selective excitation of imino/aromatic 1H can be extended to achieve multiple bond correlations that can be used to measure other 13C/15N chemical shifts in the nucleobase. As a proof of concept, we implemented two-bond 15N-1H and three-bond 13C-1H long-range correlation experiments. The compromised delays (d2) for the coherence transfer in these experiments were optimized to obtain maximum signal intensity accounting for transverse relaxation losses that occur during the generation of two-spin coherence. As the sensitivity boost due to L-optimization is determined by the excited spin, the SNt gain for the extended experiments, in reference with their respective conventional congener, scale according to their corresponding SOFAST-HMQC experiment. Since both these experiments eliminate the necessity of the intermediate spin to be isotopically enriched, they become readily amenable towards acquiring NMR data using natural abundance samples. This can be particularly important when working with unconventional chemically modified nucleobases that cannot readily be isotopically enriched (Hansen et al. 2009).
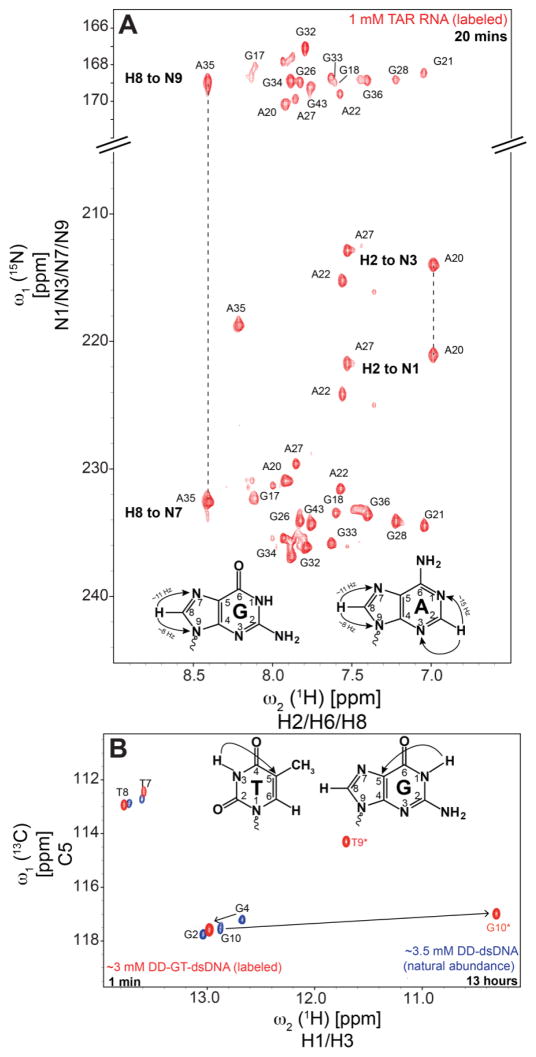
The two-bond [15N, 1H] SOFAST-HMQC experiment (Fig 1B) correlates aromatic H2 and H8 protons with N1/N3 and N7/N9, respectively, of the purine bases that are conventionally measured using an HSQC experiment (Sklenár et al. 1994; Fürtig et al. 2003). The application of this experiment is demonstrated for a 1 mM uniformly 13C/15N labeled TAR sample (Fig 3A) yielding the long range 2D [15N, 1H] spectra (SN ~16) acquired with a measurement time of 20 minutes (Table S1). The second experiment correlates the imino proton of G (H1) and T/U (H3) to C5 via the 3JCH (Fig 1A with the 15N 180° refocusing pulse at 235 ppm to refocus C5-N7 scalar coupling), which is conventionally acquired with a 2D H(NC)C experiment (Fürtig et al. 2003). The C5 chemical shift could be a valuable probe for studying structure and dynamics. As an example, the G-T wobble mispair of DD-GT-dsDNA displays a downfield shift of the T(C5), while the G(C5) a moderate upfield shift relative to canonical Watson-Crick A-T and G-C base-pairs in DD-dsDNA (Fig 3B). The long-range [13C, 1H] data for the chemical shift comparison was acquired for a 3.0 mM uniformly 13C/15N labeled sample for DD-GT-dsDNA (SN ~20) in 1 minute and for ~3.5 mM natural abundance DD-dsDNA (SN ~6) in 13 hours. As predicted, an average S/Nt gain of 1.6 and 1.9 were obtained for the long range [15N, 1H] and [13C, 1H] SOFAST-HMQC in comparison to the conventional experiment.
Fig 3.
Application of SOFAST-HMQC methodology to acquire long-range heteronuclear chemical shifts. (A) The chemical shift correlations between aromatic protons (H2/H8) with two-bond neighboring 15N in A (H2 to N1/N3 and H8 to N7/N9) and G (H8 to N7/N9) residues. Data recorded for 1 mM uniformly 13C/15N labeled TAR RNA with a measurement time of 20 minutes. (B) The 3-bond chemical shift correlation of imino protons H1/H3 with C5 of T/G for 3.5 mM natural isotopic abundance DD-dsDNA and 3.0 mM uniformly 13C/15N labeled DD-GT-dsDNA. In comparison to the canonical A–T and G–C Watson-Crick base-pairs, the T(C5) and G(C5) of the G–T mispair show a significant downfield and upfield shift, respectively.
In conclusion, we have extended prior studies showing sensitivity gains afforded by L-optimization in measurement of aromatic 13C-1H RDCs to demonstrate applications of SOFAST-HMQC geared towards recording 2D heteronuclear data and real-time applications, providing 1.4–1.7 fold sensitivity gains. We have also demonstrated that L-optimization methodology can be extended to acquire long-range heteronuclear chemical shift data that are otherwise less sensitive. Additional studies are needed to quantitatively compare the sensitivity boost yielded by SOFAST-HMQC versus BEST-TROSY (Ying et al. 2011) at various magnetic field strengths, as the sensitivity enhancements afforded by these experiments would vary upon motions experienced by the spin and the optimal experiment will also be dependent on the magnetic field strength. The L-optimized aromatic 13C-1H SOFAST-HMQC and BEST-TROSY (Ying et al. 2011) experiments provide a effective approach for acquiring multidimensional NMR data rapidly and the sensitivity boost will prove valuable in concentration limited conditions, such as ligand screening, in vivo applications (Hänsel et al. 2009) and studies involving chemically modified nucleic acids that are challenging to isotopically enrich.
Supplementary Material
Acknowledgments
This work was supported by the US National Institute for General Medical Sciences (GM103297), National Institutes of Health (NIAID R01AI066975) and an Agilent Thought Leader Award. We thank Dr. Sven G. Hyberts for his discussions on sparse sampling approaches and reconstructing the reduced 2D NMR dataset. We thank Dr. Vivekanandan Subramanian and the Duke NMR center for maintenance and support of the NMR facilities.
References
- Atreya HS, Sathyamoorthy B, Jaipuria G, Beaumont V, Varani G, Szyperski T. GFT projection NMR for efficient 1H/13C sugar spin system identification in nucleic acids. J Biomol NMR. 2012;54:337–342. doi: 10.1007/s10858-012-9687-5. [DOI] [PubMed] [Google Scholar]
- Atreya HS, Szyperski T. Rapid NMR Data Collection. Methods Enzymol. 2005;394:78–108. doi: 10.1016/S0076-6879(05)94004-4. [DOI] [PubMed] [Google Scholar]
- Barton S, Heng X, Johnson BA, Summers MF. Database proton NMR chemical shifts for RNA signal assignment and validation. J Biomol NMR. 2013;55:33–46. doi: 10.1007/s10858-012-9683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüschweiler R, Zhang F. Covariance nuclear magnetic resonance spectroscopy. J Chem Phys. 2004;120:5253–5260. doi: 10.1063/1.1647054. [DOI] [PubMed] [Google Scholar]
- Buck J, Fürtig B, Noeske J, Wöhnert J, Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc Natl Acad Sci U S A. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck J, Li Y-L, Richter C, Vergne J, Maurel M-C, Schwalbe H. NMR Spectroscopic Characterization of the Adenine-Dependent Hairpin Ribozyme. Chem BioChem. 2009;10:2100–2110. doi: 10.1002/cbic.200900196. [DOI] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR Spectroscopy. Academic Press; San Diego: 2006. [Google Scholar]
- Cromsigt JAMTC, Hilbers CW, Wijmenga SS. Prediction of proton chemical shifts in RNA – Their use in structure refinement and validation. J Biomol NMR. 2001;21:11–29. doi: 10.1023/A:1011914132531. [DOI] [PubMed] [Google Scholar]
- Dayie K. Resolution enhanced homonuclear carbon decoupled triple resonance experiments for unambiguous RNA structural characterization. J Biomol NMR. 2005;32:129–139. doi: 10.1007/s10858-005-5093-6. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMR Pipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Deschamps M, Campbell ID. Cooling overall spin temperature: Protein NMR experiments optimized for longitudinal relaxation effects. J Magn Reson. 2006;178:206–211. doi: 10.1016/j.jmr.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghbalnia HR, Bahrami A, Tonelli M, Hallenga K, Markley JL. High-Resolution Iterative Frequency Identification for NMR as a General Strategy for Multidimensional Data Collection. J Am Chem Soc. 2005;127:12528–12536. doi: 10.1021/ja052120i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RR, Bodenhausen G, Wokaun A. Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Oxford University Press; London/New York: 1987. [Google Scholar]
- Farjon J, Boisbouvier J, Schanda P, Pardi A, Simorre J-P, Brutscher B. Longitudinal-Relaxation-Enhanced NMR Experiments for the Study of Nucleic Acids in Solution. J Am Chem Soc. 2009;131:8571–8577. doi: 10.1021/ja901633y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felli IC, Brutscher B. Recent Advances in Solution NMR: Fast Methods and Heteronuclear Direct Detection. Chem Phys Chem. 2009;10:1356–1368. doi: 10.1002/cphc.200900133. [DOI] [PubMed] [Google Scholar]
- Frank AT, Bae S-H, Stelzer AC. Prediction of RNA 1H and 13C Chemical Shifts: A Structure Based Approach. J Phys Chem B. 2013;117:13497–13506. doi: 10.1021/jp407254m. [DOI] [PubMed] [Google Scholar]
- Frank AT, Horowitz S, Andricioaei I, Al-Hashimi HM. Utility of 1H NMR Chemical Shifts in Determining RNA Structure and Dynamics. J Phys Chem B. 2013;117:2045–2052. doi: 10.1021/jp310863c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman L, Scherf T, Lupulescu A. The acquisition of multidimensional NMR spectra within a single scan. Proc Natl Acad Sci U S A. 2002;99:15858–15862. doi: 10.1073/pnas.252644399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürtig B, Richter C, Wöhnert J, Schwalbe H. NMR Spectroscopy of RNA. Chem BioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- Gallego J, Varani G. Targeting RNA with Small-Molecule Drugs: Therapeutic Promise and Chemical Challenges. Acc Chem Res. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- Geen H, Freeman R. Band-selective radiofrequency pulses. J Magn Reson. 1991;93:93–141. doi: 10.1016/0022-2364(91)90034-Q. [DOI] [Google Scholar]
- Giraud N, Beguin L, Courtieu J, Merlet D. Nuclear Magnetic Resonance Using a Spatial Frequency Encoding: Application to J-Edited Spectroscopy along the Sample. Angew Chem Int Ed. 2010;49:3481–3484. doi: 10.1002/anie.200907103. [DOI] [PubMed] [Google Scholar]
- Hänsel R, Foldynová-Trantírková S, Löhr F, Buck J, Bongartz E, Bamberg E, Schwalbe H, Dötsch V, Trantírek L. Evaluation of Parameters Critical for Observing Nucleic Acids Inside Living Xenopus laevis Oocytes by In-Cell NMR Spectroscopy. J Am Chem Soc. 2009;131:15761–15768. doi: 10.1021/ja9052027. [DOI] [PubMed] [Google Scholar]
- Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM. Extending the Range of Microsecond-to-Millisecond Chemical Exchange Detected in Labeled and Unlabeled Nucleic Acids by Selective Carbon R1ρ NMR Spectroscopy. J Am Chem Soc. 2009;131:3818–3819. doi: 10.1021/ja8091399. [DOI] [PubMed] [Google Scholar]
- Hiller S, Fiorito F, Wüthrich K, Wider G. Automated projection spectroscopy (APSY) Proc Natl Acad Sci U S A. 2005;102:10876–10881. doi: 10.1073/pnas.0504818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J Biomol NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Szyperski T. GFT NMR, a New Approach To Rapidly Obtain Precise High-Dimensional NMR Spectral Information. J Am Chem Soc. 2003;125:1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
- Kovacs H, Moskau D, Spraul M. Cryogenically cooled probes a leap in NMR technology. Prog Nucl Magn Reson Spectrosc. 2005;46:131–155. doi: 10.1016/j.pnmrs.2005.03.001. [DOI] [Google Scholar]
- Krähenbühl B, Hofmann D, Maris C, Wider G. Sugar-to-base correlation in nucleic acids with a 5D APSY-HCNCH or two 3D APSY-HCN experiments. J Biomol NMR. 2012;52:141–150. doi: 10.1007/s10858-011-9588-z. [DOI] [PubMed] [Google Scholar]
- Kupče Ē, Freeman R. Wideband Excitation with Polychromatic Pulses. J Magn Reson, Ser A. 1994;108:268–273. doi: 10.1006/jmra.1994.1123. [DOI] [Google Scholar]
- Kupče Ē, Freeman R. Optimized Adiabatic Pulses for Wideband Spin Inversion. J Magn Reson, Ser A. 1996;118:299–303. doi: 10.1006/jmra.1996.0042. [DOI] [Google Scholar]
- Kupče Ē, Freeman R. Fast multi-dimensional Hadamard spectroscopy. J Magn Reson. 2003;163:56–63. doi: 10.1016/S1090-7807(03)00036-3. [DOI] [PubMed] [Google Scholar]
- Kupče Ē, Freeman R. Projection–Reconstruction Technique for Speeding up Multidimensional NMR Spectroscopy. J Am Chem Soc. 2004;126:6429–6440. doi: 10.1021/ja049432q. [DOI] [PubMed] [Google Scholar]
- Lee M-K, Gal M, Frydman L, Varani G. Real-time multidimensional NMR follows RNA folding with second resolution. Proc Natl Acad Sci U S A. 2010;107:9192–9197. doi: 10.1073/pnas.1001195107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M-H, Wang Z-F, Kuo MH-J, Hsu S-TD, Chang T-C. Unfolding Kinetics of Human Telomeric G-Quadruplexes Studied by NMR Spectroscopy. J Phys Chem B. 2014;118:931–936. doi: 10.1021/jp410034d. [DOI] [PubMed] [Google Scholar]
- Lieblein AL, Buck J, Schlepckow K, Fürtig B, Schwalbe H. Time-Resolved NMR Spectroscopic Studies of DNA i-Motif Folding Reveal Kinetic Partitioning. Angew Chem Int Ed. 2012;51:250–253. doi: 10.1002/anie.201104938. [DOI] [PubMed] [Google Scholar]
- Maciejewski MW, Fenwick M, Schuyler AD, Stern AS, Gorbatyuk V, Hoch JC. Random phase detection in multidimensional NMR. Proc Natl Acad Sci U S A. 2011;108:16640–16644. doi: 10.1073/pnas.1103723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoharan V, Fürtig B, Jäschke A, Schwalbe H. Metal-Induced Folding of Diels–Alderase Ribozymes Studied by Static and Time-Resolved NMR Spectroscopy. J Am Chem Soc. 2009;131:6261–6270. doi: 10.1021/ja900244x. [DOI] [PubMed] [Google Scholar]
- Mayzel M, Rosenlöw J, Isaksson L, Orekhov V. Time-resolved multidimensional NMR with non-uniform sampling. J Biomol NMR. 2014;58:129–139. doi: 10.1007/s10858-013-9811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishkovsky M, Frydman L. Principles and Progress in Ultrafast Multidimensional Nuclear Magnetic Resonance. Annu Rev Phys Chem. 2009;60:429–448. doi: 10.1146/annurev.physchem.040808.090420. [DOI] [PubMed] [Google Scholar]
- Mobli M, Maciejewski MW, Schuyler AD, Stern AS, Hoch JC. Sparse sampling methods in multidimensional NMR. Phys Chem Chem Phys. 2012;14:10835–10843. doi: 10.1039/C2CP40174F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish DM, Szyperski T. Simultaneously Cycled NMR Spectroscopy. J Am Chem Soc. 2008;130:4925–4933. doi: 10.1021/ja711454e. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Vogel B, Eletsky A. Longitudinal 1H Relaxation Optimization in TROSY NMR Spectroscopy. J Am Chem Soc. 2002;124:12898–12902. doi: 10.1021/ja027149q. [DOI] [PubMed] [Google Scholar]
- Piotto M, Saudek V, Sklenář V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Reining A, Nozinovic S, Schlepckow K, Buhr F, Furtig B, Schwalbe H. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature. 2013;499:355–359. doi: 10.1038/nature12378. [DOI] [PubMed] [Google Scholar]
- Rennella E, Brutscher B. Fast Real-Time NMR Methods for Characterizing Short-Lived Molecular States. Chem Phys Chem. 2013;14:3059–3070. doi: 10.1002/cphc.201300339. [DOI] [PubMed] [Google Scholar]
- Ross A, Salzmann M, Senn H. Fast-HMQC using Ernst angle pulses: An efficient tool for screening of ligand binding to target proteins. J Biomol NMR. 1997;10:389–396. doi: 10.1023/A:1018361214472. [DOI] [PubMed] [Google Scholar]
- Rovnyak D, Frueh DP, Sastry M, Sun Z-YJ, Stern AS, Hoch JC, Wagner G. Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J Magn Reson. 2004;170:15–21. doi: 10.1016/j.jmr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Sahakyan AB, Vendruscolo M. Analysis of the Contributions of Ring Current and Electric Field Effects to the Chemical Shifts of RNA Bases. J Phys Chem B. 2013;117:1989–1998. doi: 10.1021/jp3057306. [DOI] [PubMed] [Google Scholar]
- Sathyamoorthy B, Parish DM, Montelione GT, Xiao R, Szyperski T. Spatially Selective Heteronuclear Multiple-Quantum Coherence Spectroscopy for Biomolecular NMR Studies. Chem Phys Chem. 2014;15:1872–1879. doi: 10.1002/cphc.201301232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanda P, Brutscher B. Very Fast Two-Dimensional NMR Spectroscopy for Real-Time Investigation of Dynamic Events in Proteins on the Time Scale of Seconds. J Am Chem Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- Schanda P, Kupče Ē, Brutscher B. SOFAST-HMQC Experiments for Recording Two-dimensional Deteronuclear Correlation Spectra of Proteins within a Few Seconds. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- Schwalbe H, Buck J, Fürtig B, Noeske J, Wöhnert J. Structures of RNA Switches: Insight into Molecular Recognition and Tertiary Structure. Angew Chem Int Ed. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- Shaka AJ, Barker PB, Freeman R. Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson. 1985;64:547–552. doi: 10.1016/0022-2364(85)90122-2. [DOI] [Google Scholar]
- Sklenár V, Peterson R, Rejante M, Feigon J. Correlation of nucleotide base and sugar protons in a 15N-labeled HIV-1 RNA oligonucleotide by 1H-15N HSQC experiments. J Biomol NMR. 1994;4:117–122. doi: 10.1007/BF00178339. [DOI] [PubMed] [Google Scholar]
- Sripakdeevong P, Cevec M, Chang AT, Erat MC, Ziegeler M, Zhao Q, Fox GE, Gao X, Kennedy SD, Kierzek R, Nikonowicz EP, Schwalbe H, Sigel RKO, Turner DH, Das R. Structure determination of noncanonical RNA motifs guided by 1H NMR chemical shifts. Nat Meth. 2014;11 doi: 10.1038/nmeth.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat Chem Biol. 2011;7:553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyperski T, Wider G, Bushweller JH, Wüthrich K. Reduced dimensionality in triple-resonance NMR experiments. J Am Chem Soc. 1993;115:9307–9308. doi: 10.1021/ja00073a064. [DOI] [Google Scholar]
- Szyperski T, Yeh DC, Sukumaran DK, Moseley HNB, Montelione GT. Reduced-dimensionality NMR spectroscopy for high-throughput protein resonance assignment. Proc Natl Acad Sci U S A. 2002;99:8009–8014. doi: 10.1073/pnas.122224599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal A, Frydman L. Single-scan multidimensional magnetic resonance. Prog Nucl Magn Reson Spectrosc. 2010;57:241–292. doi: 10.1016/j.pnmrs.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Vega-Vazquez M, Cobas JC, Martin-Pastor M. Fast multidimensional localized parallel NMR spectroscopy for the analysis of samples. Magn Reson Chem. 2010;48:749–752. doi: 10.1002/mrc.2659. [DOI] [PubMed] [Google Scholar]
- Wenter P, Fürtig B, Hainard A, Schwalbe H, Pitsch S. Kinetics of Photoinduced RNA Refolding by Real-Time NMR Spectroscopy. Angew Chem Int Ed. 2005;44:2600–2603. doi: 10.1002/anie.200462724. [DOI] [PubMed] [Google Scholar]
- Werf RM, Tessari M, Wijmenga SS. Nucleic acid helix structure determination from NMR proton chemical shifts. J Biomol NMR. 2013;56:95–112. doi: 10.1007/s10858-013-9725-y. [DOI] [PubMed] [Google Scholar]
- Wijmenga SS, Kruithof M, Hilbers CW. Analysis of 1H chemical shifts in DNA: Assessment of the reliability of 1H chemical shift calculations for use in structure refinement. J Biomol NMR. 1997;10:337–350. doi: 10.1023/A:1018348123074. [DOI] [PubMed] [Google Scholar]
- Xu X-P, Case DA. Automated prediction of 15N, 13Cα, 13Cβ and 13C′ chemical shifts in proteins using a density functional database. J Biomol NMR. 2001;21:321–333. doi: 10.1023/A:1013324104681. [DOI] [PubMed] [Google Scholar]
- Ying J, Wang J, Grishaev A, Yu P, Wang Y-X, Bax A. Measurement of 1H–15N and 1H–13C residual dipolar couplings in nucleic acids from TROSY intensities. J Biomol NMR. 2011;51:89–103. doi: 10.1007/s10858-011-9544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.