ABSTRACT
Enteroviruses require autophagy to facilitate the formation of autophagosome (AP)-like double-membrane vesicles that provide the scaffolding for RNA replication. Here, we identify bactericidal/permeability-increasing protein (BPI) fold-containing family B, member 3 (BPIFB3) as a gene whose silencing greatly enhances coxsackievirus B (CVB) replication and induces dramatic alterations in the morphology of CVB-induced replication organelles. We show that BPIFB3 is associated with the endoplasmic reticulum (ER), and its silencing by RNA interference enhances basal levels of autophagy and promotes increased autophagy during CVB replication. Conversely, overexpression of BPIFB3 inhibits CVB replication, dramatically alters the morphology of LC3B-positive vesicles, and suppresses autophagy in response to rapamaycin. In addition, we found that, whereas silencing of core autophagy components associated with the initiation of APs in control cells suppressed CVB replication, silencing of these same components had no effect on CVB-induced autophagy or viral replication in cells transfected with BPIFB3 small interfering RNA. Based on these results, taken together, this study reports on a previously uncharacterized regulator of enterovirus infection that controls replication through a noncanonical pathway independent from the core autophagy initiation machinery.
IMPORTANCE
Coxsackievirus B (CVB) infections are commonly associated with dilated cardiomyopathy, a condition that accounts for nearly half of all heart transplants annually. During infection, CVB co-opts a cellular pathway, termed autophagy, to provide the membranes necessary for its replication. Autophagy is an evolutionarily conserved process by which cells ingest damaged organelles as a means of maintaining cell homeostasis. Here, we report on a novel regulator of autophagy, bactericidal/permeability-increasing protein (BPI) fold-containing family B, member 3 (BPIFB3), whose expression functions to restrict CVB replication by suppressing key steps in the authophagic process. We show that loss of BPIFB3 expression greatly enhances CVB replication while having no effect on replication of poliovirus, a closely related virus. Our results thus identify a novel host cell therapeutic target whose function could be targeted to alter CVB replication.
INTRODUCTION
An obligate step in the life cycle of positive-sense RNA viruses is the formation of membrane-enriched organelles that provide the structural support for viral replication; these are termed replication organelles. Multiple mechanisms have been proposed for the generation of these membranes, including manipulation of the host autophagic pathway, a process that removes damaged organelles via the formation of double-membrane-bound vesicles. Despite the strong association between RNA viruses and autophagy, many of the host cell components that regulate virus-host membrane manipulation remain poorly defined.
There are at least three autophagic pathways—macroautophagy (the most common form), microautophagy, and chaperone-mediated autophagy. These distinct pathways differ in their morphological characteristics (such as the specific appearance of double-membrane vesicles formed during macroautophagy), the organisms in which they exist (macroautophagy and microautophagy occur in multi- and unicellular organisms, whereas chaperone-mediated autophagy has thus far been determined to be restricted to mammalian cells), and the specific cellular components regulating each pathway. Macroautophagy (here referred to as autophagy) is initiated by the formation of an isolation membrane (also termed the phagophore) to form the characteristic double-membrane vesicle—termed the autophagosome (AP). APs can then fuse with endosomes to form amphisomes or fuse with lysosomes to form autolysosomes, which expose the material within the APs to lysosomal hydrolases, resulting in their degradation, a process referred to as autophagic flux. Several cellular organelles provide membranes for the isolation membrane, including the endoplasmic reticulum (ER) (1, 2), Golgi complex (3), ER-Golgi complex intermediate compartment (ERGIC) (4), the mitochondrial outer membrane (5), and the mitochondrial-associated membrane (MAM) at ER-mitochondria contacts (6). The process of autophagy is exquisitely controlled and orchestrated by a growing list of cellular factors that tightly regulate various aspects of the autophagic pathway. These components regulate specific aspects of the autophagy pathway, including autophagy induction (e.g., ULK1/2, ATG13, and FIP200), nucleation of the phagophore (e.g., ATG9, ATG14, beclin-1, and UVRAG), and elongation (e.g., ATG7, the LC3 ubiquitin-like [Ubl] conjugation system, and the ATG12-ATG5-ATG16L1 Ubl conjugation system) (reviewed in references 7 and 8). More recently, specific regulators of autophagic fusion have been identified and include the N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) syntaxin-17, vesicle-associated membrane protein 8 (VAMP8), and soluble NSF attachment protein 29 (SNAP29) (9, 10). However, there are cases in which autophagy proceeds without the “core” autophagic machinery, as is the case in beclin-1-independent forms of autophagy (11, 12), in response to cytotoxic stressors (13), and the specific clearance of the ER by autophagy (reticulophagy) in response to ER stress/expansion (14).
Enteroviruses such as coxsackievirus B (CVB) and poliovirus (PV) induce the formation of AP-like double-membrane vesicles during their replication (15–19). CVB replication relies on expression of core autophagy components, and silencing or depletion of these components in vitro (18) or in vivo (19) suppresses viral replication. Although CVB and PV share a requirement for autophagy to support their replication, their effects on AP-lysosome fusion in completion of their infectious life cycles may differ. Whereas PV infection induces AP-lysosome fusion, a step thought to be required for virion maturation (20), studies have suggested that CVB inhibits autolyosome formation (18). This inhibition was suggested to be a mechanism by which the virus evades lysosomal degradation, given that silencing of a lysosomal SNARE (VAMP2) enhanced CVB replication (18). However, as VAMP2 has not been implicated in AP-lysosome fusion and more recent studies have identified the specific lysosomal SNAREs associated with this process (VAMP8 and SNAP29) (9, 10), the role of AP-lysosome fusion in the replication of CVB remains unclear.
To identify host cell factors that regulate enterovirus replication, we previously conducted comparative high-throughput genome-scale RNA interference (RNAi) screening using CVB and PV (21). In the current study, we report a host cell component first identified by this primary RNAi screen that regulates autophagy to specifically promote CVB, but not PV, replication. This component—bactericidal/permeability-increasing protein (BPI) fold-containing family B, member 3 (BPIFB3, also known as long palate, lung, and nasal epithelium clone [LPLUNC3])—is a largely uncharacterized protein of the BPI/lipopolysaccharide-binding protein (LBP) family of antibacterial components (22). Members of the BPI/LBP family share significant sequence homology with cholesterol ester transfer protein (CETP) and phospholipid transfer protein (PLTP), both of which are involved in lipid transport in blood plasma (23). BPIFB3 has not been functionally characterized, and any assigned putative function is largely due to its homology with lipid-binding antimicrobial proteins of the LBP/BPI superfamily. In the current study, we show that BPIFB3 localizes to the ER, and its RNAi-mediated silencing induces significant enhancements in CVB, but not PV, replication and dramatic alterations in the morphology of CVB-induced replication organelles. In addition, we show that silencing of BPIFB3 enhances basal levels of autophagy and promotes enhanced autophagy during CVB replication, whereas its overexpression leads to alterations in LC3B localization and inhibits autophagy. Surprisingly, we found that, whereas silencing of core components of the autophagy initiation machinery in control cells suppressed CVB replication, silencing of these components had no effect on CVB-induced autophagy or on viral replication in cells depleted of BPIFB3. Taking together these results, we were able to identify a previously uncharacterized regulator of enterovirus infection that controls replication in a virus-specific manner through the facilitation of autophagy via a noncanonical pathway independent from the core autophagic initiation machinery.
RESULTS
Identification of BPIFB3 as a regulator of CVB, but not PV, replication.
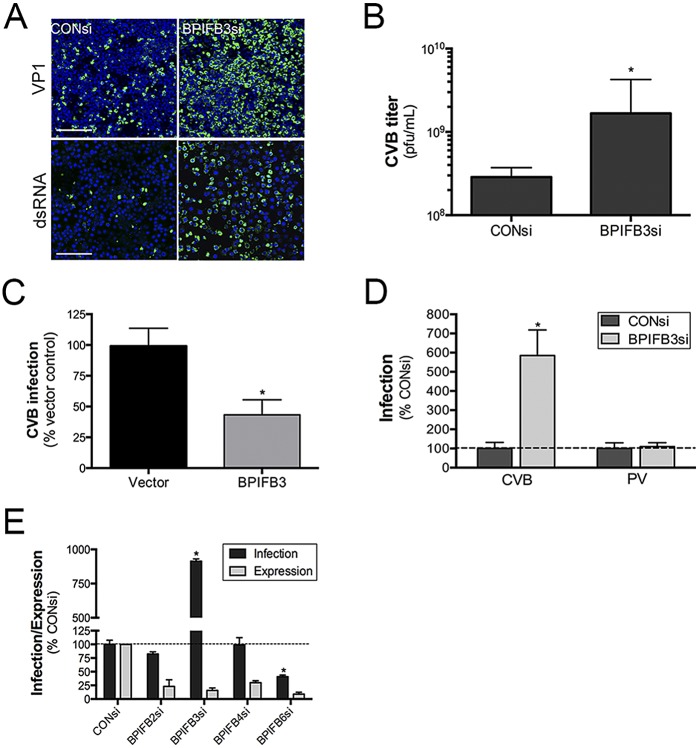
We previously identified BPIFB3 by RNAi screening as a gene whose depletion significantly enhanced the replication of CVB in human brain microvascular endothelial cells (HBMEC), an in vitro model of the blood-brain barrier (21). As our initial screening was conducted using pooled small interfering RNAs (siRNAs), we tested the effects of individual siRNAs on CVB infection and found that two siRNAs against BPIFB3 (BPIFB3si-1 and BPIFB3si-3) exerted proviral effects and induced a corresponding knockdown of BPIFB3 expression, with BPIFB3si-1 exhibiting the greatest effects (Fig. 1A; see also Fig. S1 in the supplemental material). This siRNA was chosen for all subsequent experiments. The enhancement of CVB infection by BPIFB3 silencing correlated with an ~1-log enhancement in CVB titers (Fig. 1B). We also found that CVB replication was suppressed in cells overexpressing BPIFB3 (Fig. 1C). Unlike CVB, we found that depletion of BPIFB3 had no effect on PV replication (Fig. 1D) and did not impact PV titers (data not shown). In addition, we found that the enhancement of CVB replication was specific to BPIFB3, and silencing of other members of the BPI-containing family (including BPIFB2, BPIFB4, and BPIFB6) did not enhance replication (Fig. 1E).
FIG 1 .
CVB replication is enhanced by BPIFB3 silencing. (A) Enhanced CVB infection in HBMEC transfected with BPIFB3-1 siRNA, compared to CONsi-transfected cells. (Top row) VP1 staining is shown in green. (Bottom row) dsRNA is stained green, and DAPI-stained nuclei are shown in blue. Bar, 100 µm. (B) CVB titers from HBMEC transfected with CONsi or BPIFB3si, determined by plaque assay. (C) CVB infection in U2OS cells transfected with vector or BPIFB3-Flag and infected with CVB (5 PFU/cell) for ~16 h. Data shown are the percent infection (as assessed by immunofluorescence microscopy), normalized to levels for vector-transfected controls. (D) HBMEC transfected with control (CONsi) or BPIFB3 (BPIFB3si) siRNAs were infected with PV (1 PFU/cell) or CVB (1 PFU/cell) for ~18 h, and infection was assessed by RT-qPCR. (E) CVB infection was assessed by RT-qPCR in HBMEC transfected with the indicated BPIFB family member siRNAs or CONsi. Data in panels B to E are means ± standard deviations. *, P < 0.01.
BPIFB3 is localized to the ER.
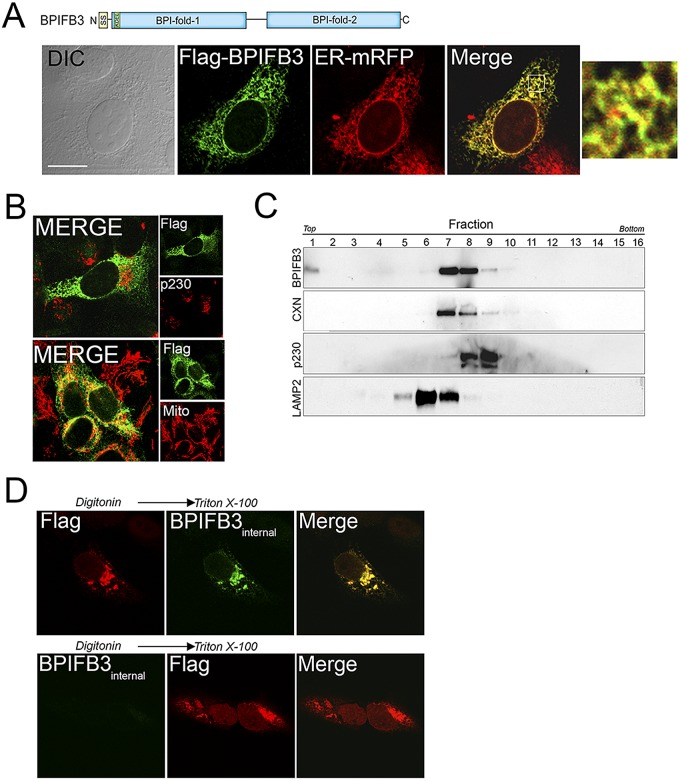
The subcellular localization of BPIFB3 is unknown, although Rya3, the rat homolog of BPIFB3, has been proposed to localize diffusely in the cytoplasm (24). To determine the subcellular localization of BPIFB3 in human cells, we constructed a C-terminal Flag fusion construct (a schematic of BPIFB3-Flag is shown at the top of Fig. 2A). We found that BPIFB3-Flag localized exclusively to the ER (Fig. 2A). In contrast, BPIFB3 did not localize with markers of mitochondria, the Golgi complex, early endosomes, or lipid droplets (Fig. 2B; see also Fig. S2A in the supplemental material). To confirm its ER localization, we performed subcellular fractionation studies in human osteosarcoma U2OS cells stably expressing BPIFB3-Flag and found that BPIFB3 was distributed to fractions enriched in the ER-associated marker calnexin (Fig. 2C; see also Fig. S2B). BPIFB3 contains a KDEL sequence at the N terminus that is located in close proximity to the signal sequence. However, mutation of this sequence to AAEL, a sequence shown previously to abolish KDEL receptor binding (25), had no effect on its ER targeting (see Fig. S2C).
FIG 2 .
Localization of BPIFB3 to the ER. (A, top) Schematic of BPIFB3. (Bottom) Confocal microscopy for Flag (green) and ER-mRFP (red) in U2OS cells transfected with BPIFB3-Flag and infected with CellLights ER-RFP baculovirus. (Left) Differential interference contrast (DIC) image. (Right) A 5× magnification of the area indicated by the white box in the merged image. (B) Confocal microscopy for Flag (green) and either p230/Golgi (red, top) or MTCO2 (to label mitochondria; red, bottom) in U2OS cells transfected with BPIFB3-Flag at ~48 h posttransfection. (C) Subcellular fractionation of BPIFB3 in U2OS cells stably expressing BPIFB3-Flag. Shown are immunoblots from collected fractions for BPIFB3-Flag, calnexin (CXN), p230/Golgin, and LAMP2. (D) Confocal microscopy of U2OS cells transiently transfected with BPIFB3-Flag. At 48 h posttransfection, cells were permeabilized with digitonin, fixed with PFA, and then incubated with anti-Flag-M2 (top row) or BPIFB3 (bottom row) antibodies. Cells were then permeabilized with Triton X-100 and stained with anti-BPIFB3 (top row) or anti-Flag M2 (bottom row) antibodies. Bar, 10 µm.
To determine the topology of BPIFB3 within the ER, we applied a staining technique that relies on a two-step permeabilization procedure utilizing digitonin to first permeabilize the plasma membrane, followed by Triton X-100 to permeabilize intracellular membranes (26). We found that the Flag-tagged C-terminal domain of BPIFB3 (recognized by the anti-Flag antibody) was located in the cytosol and was readily detectable by digitonin permeabilization alone (Fig. 2D, top row), whereas an internal region of BPIFB3 (detected by an antibody recognizing portions of both BPI-like domains) was localized within the ER lumen and required Triton X-100 permeabilization for its detection (Fig. 2D, bottom row).
BPIFB3 silencing promotes the formation of megaphagosomes in CVB-infected cells.
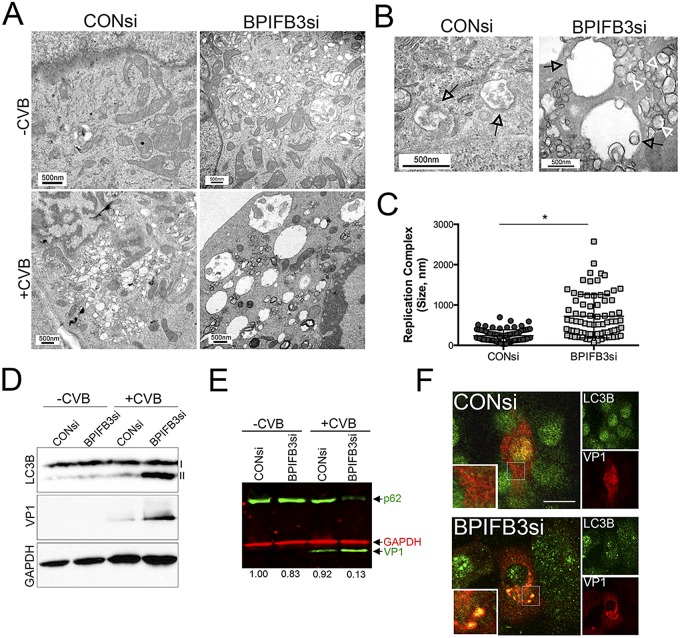
CVB infection induces the formation of double-membrane vesicles that resemble APs when observed by electron microscopy (EM) (15, 16, 27, 28). In cells transfected with control siRNA (CONsi), we found that CVB infection led to the formation of small vesicles (~100 to 300 nm) that often resembled APs, given their double membranes (Fig. 3A to C). In contrast, in cells transfected with BPIFB3si, CVB infection induced the formation of very large replication organelles that ranged in size from ~300 nm to as large as >2 µm (Fig. 3A to C). These organelles were not apparent in uninfected cells (Fig. 3A, top row) and only resulted as a consequence of CVB infection (Fig. 3A, bottom row).
FIG 3 .
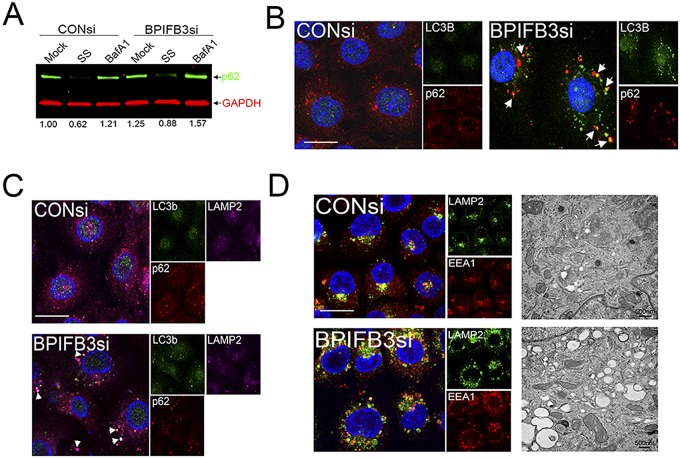
Silencing of BPIFB3 enhances autophagy during CVB infection. (A) Electron micrographs of uninfected (top row) or CVB-infected (3 PFU/cell for ~16 h) (bottom row) HBMEC transfected with CONsi (left panels) or BPIFB3si (right panels). (B) High-magnification images of CVB-induced replication organelles in HBMEC transfected with CONsi (left) or BPIFB3si (right). Black arrows denote replication organelles in CONsi-transfected cells or megaphagosomes in BPIFB3si-transfected cells. White arrows denote smaller, double-membrane-bound vesicles in BPIFB3si-transfected cells. (C) Size of CVB-induced replication organelles in cells transfected with CONsi or BPIFB3si, assessed by EM. A total of approximately 100 independent organelles from at least 20 unique cells were quantified. *, P < 0.001. (D) Immunoblots for LC3B (top), VP1 (middle), and GAPDH (bottom) in uninfected or CVB-infected HBMEC transfected with CONsi or BPIFB3si. I, nonlipidated LC3B; II, AP-associated LC3B. (E) Results of dual-color immunoblot analysis using a LI-COR Odyssey infrared imaging system and antibodies specific for p62 and VP1 (800 nm; green) and GAPDH (700 nm; red) in HBMEC transfected with CONsi or BPIFB3si and uninfected or infected with CVB (3 PFU/cell) for 24 h. Numbers at the bottom show densitometry results for the level of p62, normalized based on GAPDH levels, and presented as the fold change from uninfected CONsi-transfected cells. (F) HBMEC transfected with CONsi or BPIFB3si were infected with CVB for ~8 h and then fixed and stained for VP1 (in red) and LC3B (in green). Areas of colocalization appear in yellow. Bar, 10 µm.
In pancreatic acinar cells, CVB-induced APs are enlarged (>1 µm) and have been termed “megaphagosomes” (28). The induction of these megaphagosomes requires components associated with autophagy, such as ATG5 (28). Given that the enlarged replication organelles in BPIFB3-depleted cells shared characteristics with those previously described megaphagosomes, we assessed the impact of BPIFB3 silencing on CVB-induced autophagy. We found that silencing of BPIFB3 enhanced autophagy in CVB-infected cells, as assessed by the conversion of nonlipidated LC3B (LC3B-I) to cleaved, AP-associated, and lipidated LC3B-II (Fig. 3D). Consistent with an enhancement in autophagy, we found that CVB infection elicited the pronounced degradation of p62/SQSTM1, an LC3-binding protein that is degraded upon AP-lysosome fusion (29, 30), in BPIFB3si- but not CONsi-transfected cells (Fig. 3E). In addition, we found that BPIFB3si increased the association between LC3B and viral replication complexes, as assessed by VP1 immunofluorescence (Fig. 3F). Taken together, these data suggest that loss of BPIFB3 stimulates autophagy in CVB-infected cells to enhance viral replication.
BPIFB3 silencing enhances autophagy.
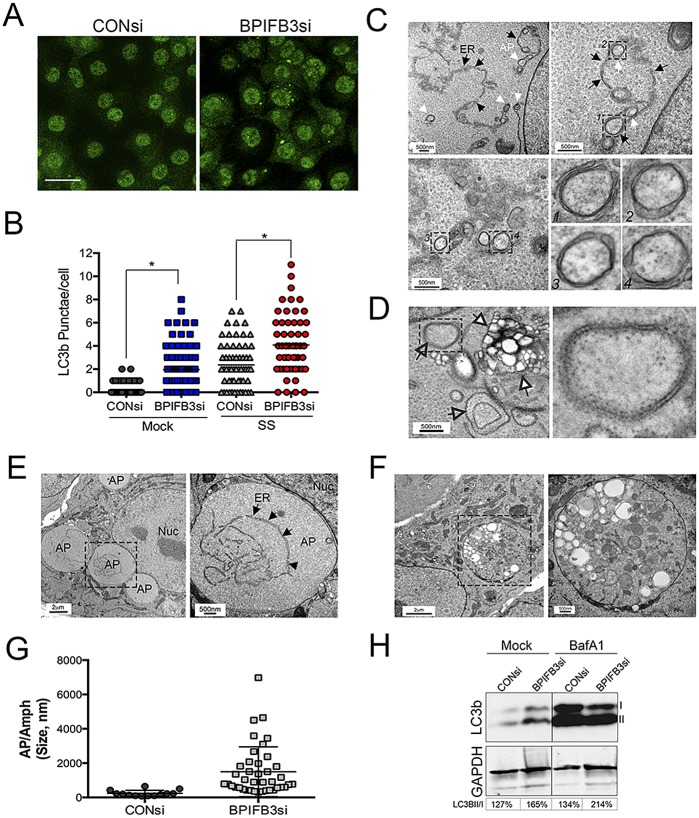
Because we found that silencing of BPIFB3 exerted dramatic effects on the morphology of CVB-induced replication organelles and enhanced autophagy during CVB replication, we next assessed the effects of BPIFB3 knockdown on autophagy, both under basal and nutrient-deprived conditions. We found that silencing of BPIFB3 led to a significant enhancement of APs, as assessed by the formation of endogenous LC3B puncta (Fig. 4A and B). We also observed an enhancement in the induction of autophagy in HeLa and human kidney 786-O cells (see Fig. S3A to C in the supplemental material), suggesting that the regulation of autophagy by BPIFB3 is not specific to endothelial cells. In addition, we found that the magnitude of LC3B punctum formation in response to serum starvation was enhanced in BPIFB3si-transfected cells compared to control cells (Fig. 4B). Unlike BPIFB3, we found that silencing of other members of the BPI fold containing family (BPIFB2, BPIFB4, and BPIFB6) had no impact on LC3B puncta, suggesting that other BPI fold-containing family members do not participate in this process (data not shown).
FIG 4 .
Depletion of BPIFB3 induces autophagy. (A) HBMEC transfected with control (CONsi) or BPIFB3 (BPIFB3si) siRNAs for ~48 h were immunostained for LC3B (green). Bar, 20 µm. (B) Quantification of the number of LC3B puncta per cell in HBMEC transfected with CONsi or BPIFB3si and either mock treated or serum starved (SS) for 3 h. (C) Electron micrographs of HBMEC transfected with CONsi or BPIFB3si for ~48 h. Black arrows denote the ER membrane, and white arrows denote autophagosomes forming along the ER membrane. Select vesicles are magnified at the bottom right and correspond to the numbered black-hatched areas shown. (D) Electron micrographs of HBMEC transfected with CONsi or BPIFB3si for ~48 h. Empty black arrows denote RER-lined autophagosomes. White arrows denote an amphisome or multivesicular body. On the right is a magnified image of the black-hatched area shown at left. (E and F) Electron micrographs of HBMEC transfected with BPIFB3si for ~48 h and treated with BafA1 for ~8 h. In panel E, black arrows denote the ER membrane within an enlarged autophagosome, shown in the black-hatched box at left. nuc, nucleus. In panel F, an enlarged amphisome is shown. At right is a magnified image of the black-hatched box on the left. (G) Quantification of the sizes of autophagosomes and amphisomes (Amph) in cells transfected with CONsi or BPIFB3si and treated with BafA1. APs/Amphs were quantified from at least 50 unique cells. (H) Immunoblotting for LC3B and GAPDH in HBMEC transfected with CONsi or BPIFB3si and treated with BafA1. I, nonlipidated LC3B; II, lipdated AP-associated LC3B. At the bottom of the panel, the percentages of LC3B-II normalized to LC3B-I are shown (determined by densitometry).
One of the earliest steps in autophagy is the formation of the isolation membrane (IM), which elongates and fuses to form the AP. The ER is one of the cellular organelles that has been implicated in providing the membranes necessary for the formation of the IM (1, 31). In cells transfected with BPIFB3 siRNA (Fig. 4C), we observed by EM the extensive formation of double-membrane-bound APs directly forming along the ER membrane, as well as fully formed APs that were derived from the rough ER (RER), a site associated with AP biogenesis (32) (Fig. 4D). The double-membrane cup-shaped structures were fully closed (and likely represented fully formed APs) and partially closed (and likely represented elongating IMs) (Fig. 4C, lower right). These APs and IMs were not observed in any cells transfected with control siRNA (data not shown). The size and presence of APs were bolstered by treatment of cells with bafilomycin A1 (BafA1), which induces the accumulation of APs by altering the activity of lysosomal hydrolases and/or by inhibiting AP-lysosome fusion (33), and led to the appearance of enlarged APs (Fig. 4E) and amphisomes (Fig. 4F). The sizes of both APs and amphisomes in BPIFB3si-transfected cells treated with BafA1 were significantly larger than those in CONsi-transfected cells (Fig. 4G). Consistent with enhanced autophagy in BPIFB3si-transfected cells, we also detected enhanced levels of LC3B-II in BPIFB3si-transfected cells under resting conditions, and these levels were increased by treatment of cells with BafA1 (Fig. 4H).
APs induced by BPIFB3 silencing are not degradative but associate with LAMP-2.
The enhanced numbers of APs in BPIFB3si-transfected cells suggested that either the extent of AP initiation was enhanced and/or the level of AP maturation/flux was inhibited. To determine if autophagic flux was inhibited by BPIFB3si under resting conditions and in response to nutrient deprivation, we assessed the levels of p62. We found that BPIFB3si transfection increased LC3B puncta in resting cells and in response to serum starvation (Fig. 4A and B), but we did not detect any enhancement in the degradation of p62 in resting cells or in serum-starved cells depleted of BPIFB3 (Fig. 5A). In addition, we found that RNAi-mediated silencing of BPIFB3 led to the association of p62 with punctate structures that were largely positive for LC3B (Fig. 5B).
FIG 5 .
Silencing of BPIFB3 does not enhance autophagosome-lysosome fusion but alters lysosomal size. (A) Dual-color immunoblot analysis using a LI-COR Odyssey infrared imaging system for p62 (800 nm; green) and GAPDH (700 nm; red) in cells transfected with CONsi or BPIFB3si and either mock or BafA1 treated. Numbers at the bottom of the panel are densitometry results for the level of p62, normalized to GAPDH, and are presented as the fold change from mock-treated CONsi-transfected cells. (B) HBMEC transfected with CONsi or BPIFB3si for ~48 h were immunostained for LC3B (green) and p62 (red). Areas of colocalization appear yellow (white arrows). DAPI-stained nuclei are shown in blue. (C) HBMEC transfected with CONsi or BPIFB3si for ~48 h were immunostained for LC3B (green), p62 (red), and LAMP2 (magenta). Areas of colocalization appear white (white arrows). DAPI-stained nuclei are shown in blue. (D, left) HBMEC were transfected with CONsi or BPIFB3si for ~48 h and immunostained for LAMP2 (green) and EEA1 (red). (Right) EM image of enlarged endosomes in cells transfected with CONsi or BPIFB3si. Bars, 10 µm.
The late endosomal and lysosome-associated membrane protein 2 (LAMP2) is involved in AP maturation, and LAMP2 deficiency and/or altered function induces the accumulation of APs (34). We found that many of the LC3B-positive puncta induced by BPIFB3 depletion were positive for LAMP2 and contained p62 (Fig. 5C), suggesting that the APs formed by silencing of BPIFB3 are capable of fusing with late endosomes and/or lysosomes but are nondegradative in nature, given the accumulation of p62. We therefore investigated the impact of silencing of BPIFB3 on early endosomes (by using early endosome antigen-1 [EEA1]), late endosomes (using Rab7), and lysosomes (using LAMP2). We found that transfection of cells with BPIFB3si had a dramatic effect on the endosomal pathway and led to a significant enhancement in the size of early (EEA1-posiitve) and late (Rab7-positive) endosomes and lysosomes (LAMP2 positive) (Fig. 5D; see also Fig. S4A in the supplemental material). However, the total numbers of endosomes and lysosomes did not change in BPIFB3si-transfected cells (see Fig. S4B). Similar results were obtained in HeLa cells (see Fig. S4C). Enlarged, early endosomes and lysosomes were also apparent by EM in BPIFB3-depleted cells treated with BafA1 to inhibit endosomal maturation (35) and lysosomal acidification (36) (Fig. 5D, right panel). These data support a model wherein silencing of BPIFB3 alters endo-lysosomal trafficking and alters AP-lysosome fusion.
Overexpression of BPIFB3 alters LC3B localization and inhibits autophagy.
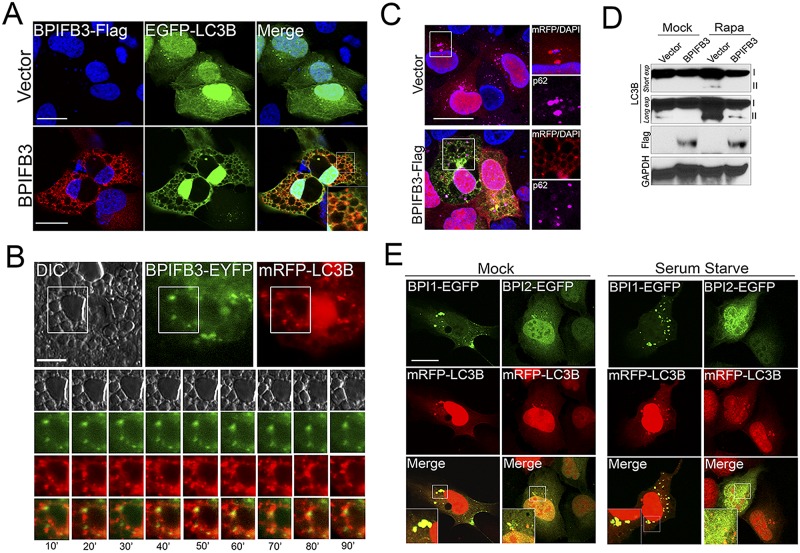
Because we found that silencing of BPIFB3 enhanced autophagy, we next determined whether its overexpression might induce an opposite effect. To do this, we cotransfected BPIFB3-Flag fusions with either enhanced green fluorescent protein (EGFP)- or monomeric red fluorescent protein (mRFP)-fused LC3B and assessed the effects of this transfection on LC3B localization and autophagy induction. As expected, EGFP-LC3B localized diffusely and/or in small punctate structures in cells cotransfected with the vector control under naive conditions (Fig. 6A). Surprisingly, we found that coexpression of EGFP-LC3B (Fig. 6A) or mRFP-LC3B (see Fig. S5A in the supplemental material) with BPIFB3-Flag led to the appearance of enlarged cytoplasmic vacuoles from which LC3B was largely excluded. This effect was specific for BPIFB3, as overexpression of V5-fused BPIFB2 or BPIFB6 had no effect on the localization of EGFP- or mRFP-LC3B (data not shown, but see Fig. S5B). Moreover, these vacuoles did not form when BPIFB3 was expressed alone or when BPIFB3 was coexpressed with other components, such as LAMP1 (see Fig. S5A and C).
FIG 6 .
Overexpression of BPIFB3 inhibits autophagy. (A) U2OS cells transfected with BPIFB3-Flag (or vector control) and EGFP-LC3B for ~48 h were fixed and immunostained for Flag (in red). DAPI-stained nuclei are shown in blue. (B) Select frames (taken at 10-min intervals) from time-lapse live-cell imaging of U2OS cells transfected with BPIFB3-EYFP and mRFP-LC3B and treated with rapamycin from ~60 min posttreatment. Note that BPIFB3-EYFP and mRFP-LC3B do not colocalize (see Movie S1 in the supplemental material for the complete movie). (C) U2OS cells transfected with BPIFB3-Flag (or vector control) and mRFP-LC3B for ~48 h were fixed and immunostained for Flag (in green) and p62 (in magenta). On the right is a magnified image of the area denoted by the white box. DAPI-stained nuclei are shown in blue. (D) Immunoblotting for LC3B, Flag, and GAPDH in U2OS cells transfected with EGFP-LC3B and BPIFB3-Flag (or vector control) and mock or rapamycin treated for ~12 h. Two exposures are included for LC3B (“short” and “long”). Nonlipidated (I) and lipdated AP-associated LC3B (II) results are shown. (E) U2OS cells transfected with either EGFP-BPI-1 or EGFP-BPI-2 and mRFP-LC3B for ~48 h and cultured under nutrient-rich (mock) or nutrient-deprived (serum starved) conditions (for 4 h). Bars, 10 µm.
To follow the localization of BPIFB3 and LC3B in vacuole-containing cells, we performed real-time imaging of enhanced yellow fluorescent protein (EYFP)-fused BPIFB3 and mRFP-LC3B in live cells under rapamycin-treated conditions. In rapamycin-treated cells, we found that the movements of LC3B puncta were restricted in cells containing BPIFB3-induced vacuoles and that BPIFB3 did not appear to colocalize with these puncta (Fig. 6B). In contrast, vector-transfected cells formed LC3B puncta normally, which moved freely throughout the cytoplasm (see Fig. S6A and Movies S1 and S2 in the supplemental material). In addition, we found a reduction in the association between mRFP-LC3B and p62 in BPIFB3-overexpressing cells, suggesting a defect in the induction and/or maturation of APs (Fig. 6C). Consistent with this, we did not observe any association between the enlarged LC3B vacuoles and LAMP2 in BPIFB3-overexpressing cells (data not shown).
In addition, we found that overexpression of BPIFB3 inhibited the induction of autophagy in rapamycin-treated cells expressing EGFP-LC3B and inhibited the conversion of LC3B-I to LC3B-II (Fig. 6D). Collectively, these data showed that overexpression of BPIFB3 induces the formation of enlarged LC3B vacuoles which are not associated with lysosomes and inhibits autophagy.
The first BPI fold of BPIFB3 localizes with LC3B puncta and induces LC3B vacuoles.
BPIFB3 consists of two large BPI folds (BPI-1 and BPI-2) (Fig. 2A, top, schematic). To determine the effects of these individual domains on LC3B localization, we expressed EGFP-fused BPI-1 and BPI-2 with mRFP-LC3B in both mock- and rapamycin-treated cells. We found that BPI-1 exhibited strong colocalization with mRFP-LC3B puncta in mock-treated cells, whereas BPI-2 exhibited little colocalization (Fig. 6E). The localizations of BPI-1 and LC3B were enhanced when cells were nutrient depleted (Fig. 6E). We also found that expression of BPI-1 alone was sufficient to induce the formation of enlarged mRFP-LC3B vacuoles under both mock treatment (data not shown) or rapamycin treatment conditions, similar to those observed in cells overexpressing full-length BPIFB3 (see Fig. S6B in the supplemental material). We did not detect any induction of mRFP-LC3B vacuoles in cells expressing BPI-2 under any conditions. In addition, we found that the individual BPI folds of BPIFB6 did not colocalize significantly with mRFP-LC3B or induce mRFP-LC3B vacuoles (data not shown). These findings implicate the first BPI fold of BPIFB3 in the regulation of autophagy and LC3B vacuoles.
Silencing of BPIFB3 enhances CVB infection independently from components of the core autophagy initiation machinery.
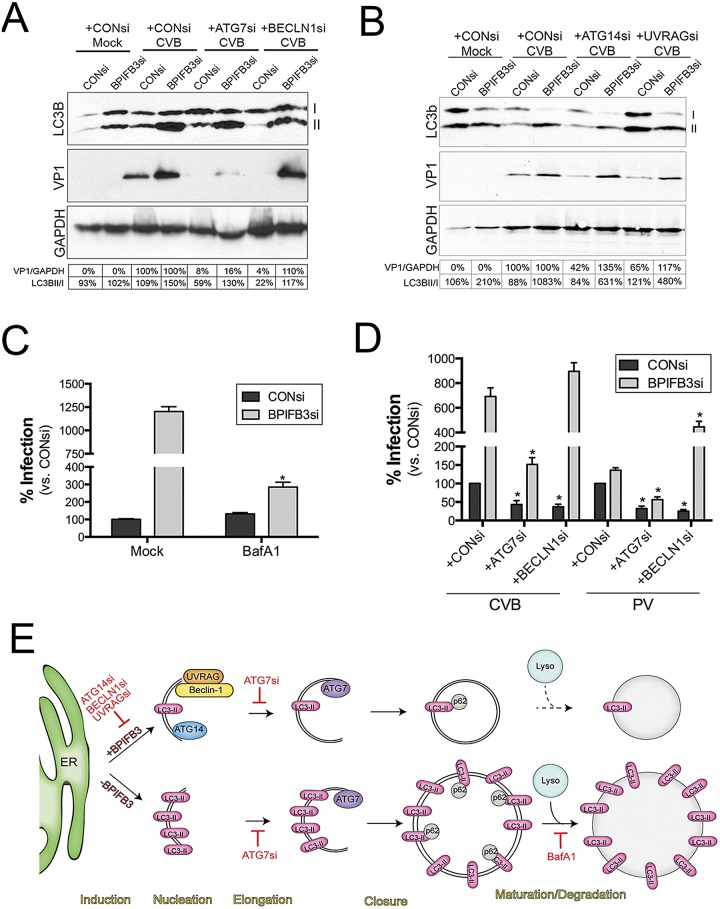
Because we found that silencing of BPIFB3 enhanced autophagy in response to CVB infection, we next assessed the impact of silencing components of the core autophagy machinery on the BPIFB3si-mediated enhancement of CVB replication. As expected, we found that silencing of ATG7, which facilitates AP elongation, and beclin-1, which regulates AP formation, potently suppressed CVB infection in cells cotransfected with CONsi (Fig. 7A). In addition, we found that silencing of ATG7 also suppressed CVB replication in cells cotransfected with BPIFB3si (Fig. 7A). However, ATG7 silencing did not inhibit the CVB-induced formation of LC3B-II in BPIFB3si-transfected cells infected with CVB (Fig. 7A), consistent with its role in elongation rather than initiation. Surprisingly, we found that silencing of beclin-1 had no effect on CVB replication in BPIFB3si-transfected cells (Fig. 7A). Moreover, the enhancement in the formation of LC3B-II in cells transfected with BPIFB3si and infected with CVB was not inhibited by beclin-1 silencing (Fig. 7A), despite beclin-1’s role in the initiation of canonical APs.
FIG 7 .
Silencing of BPIFB3 enhances CVB infection by the induction of autophagy, independent of the core autophagy initiation machinery. (A) Immunoblots for LC3B (top), VP1 (middle), and GAPDH (bottom) from HBMEC transfected with CONsi or BPIFB3si, cotransfected with either control, ATG7, or beclin-1 (BECLN1) siRNAs, and then infected with CVB (1 PFU/cell). Nonlipidated (I) and lipdated AP-associated LC3B (II) results are shown. At bottom, densitometry analysis for VP1 (normalized to GAPDH results and presented as the percent change from CONsi-transfected levels for either CONsi or BPIFB3si) and the percentage of LC3B-II normalized to LC3B-I. (B) Immunoblots for LC3B (top), VP1 (middle), and GAPDH (bottom) from HBMEC transfected with CONsi or BPIFB3si, cotransfected with either control, ATG14, of UVRAG siRNAs, and then infected with CVB (1 PFU/cell). Nonlipidated (I) and lipdated AP-associated LC3B (II) results are shown. At the bottom, densitometry analysis results for VP1 (normalized to GAPDH and presented as the percent change from CONsi-transfected levels for either CONsi or BPIFB3si) and the percentage of LC3B-II normalized to LC3B-I are shown. (C) HBMEC transfected with CONsi or BPIFB3si were infected with CVB in the absence (mock) or presence of BafA1, and then infection was assessed by RT-qPCR. Data were normalized to mock-treated conditions in CONsi-transfected cells. (D) HBMEC transfected with CONsi or BPIFB3si and either CONsi, ATG7si, or beclin-1 siRNAs were infected with PV (1 PFU/cell) or CVB (1 PFU/cell) for ~18 h, and infection was assessed by RT-qPCR. Data in panels C and D are means ± standard deviations. *, P < 0.01. (E) Schematic for the mechanism by which BPIFB3 regulates CVB replication. At top, in cells expressing BPIFB3, CVB replication induces autophagy and requires the expression/activity of core components of the autophagy initiation machinery (ATG14, UVRAG, beclin-1) and a component involved in elongation (ATG7). Hatched lines indicate that it is unknown if CVB elicits autophagic flux. Below, in cells with low levels of BPIFB3, CVB infection proceeds in the absence of the core initiation machinery, enhancing the levels of LC3B-II, promoting autophagic flux, and enhancing the sizes of replication organelles.
Because ATG7 and beclin-1 regulate different steps in autophagy (elongation versus initiation, respectively) and exerted differing effects on CVB replication in BPIFB3si-transfected cells, we tested the effects of silencing of other regulators of autophagy initiation (ATG14 and UVRAG) on CVB replication in CONsi- and BPIFB3si-transfected cells. Similar to our findings with beclin-1, we found that silencing of ATG14 and UVRAG suppressed CVB replication in CONsi-transfected but not BPIFB3si-transfected cells (Fig. 7B). In addition, the enhancement of LC3B-II formation in BPIFB3si-transfected cells infected with CVB was not inhibited by silencing of either ATG14 or UVRAG (Fig. 7B). In all experiments, efficacy of silencing was assessed by reverse transcriptase quantitative PCR (RT-qPCR) and/or immunoblotting (see Fig. S7 in the supplemental material).
For PV, the formation of autolysosomes may be required for viral maturation, and alterations in the acidification of autolysosomes by agents such as BafA1 suppress viral replication (20). Given that silencing of BPIFB3 enhanced autophagic flux in CVB-infected cells (Fig. 3E), we next assessed whether this treatment would alter the sensitivity of CVB to BafA1, which inhibits this process. Whereas CVB was unaffected by BafA1 treatment in control siRNA-transfected cells, we found that viral replication was sensitive to BafA1 in cells depleted of BPIFB3 (Fig. 7C), suggesting that the promotion of autophagic flux induced by BPIFB3si is involved in its promotion of CVB replication. Taken together, these findings suggest that silencing of BPIFB3 expression enhances CVB replication independently from components of the core autophagic machinery associated with the initiation of autophagy.
Silencing of BPIFB3 enhances PV infection in the absence of beclin-1.
Although silencing of BPIFB3 enhanced CVB replication, we found that this treatment had no significant impact on PV replication (Fig. 1D). Because we found that silencing of core initiation components of the autophagy pathway had no effect on the enhancement of CVB replication in BPIFB3-silenced cells, we assessed whether silencing of BPIFB3 in cells also depleted of ATG7 and/or beclin-1 would alter PV replication. Silencing of BPIFB3 in cells transfected with control siRNA had no significant effect on PV replication, as expected (Fig. 7D). Similar to our results with CVB, we found that silencing of ATG7 in both control and BPIFB3 siRNA-transfected cells suppressed PV replication (Fig. 7D). In CONsi-transfecetd cells, we also found that silencing of beclin-1 inhibited PV replication (Fig. 7D). In contrast, we found that silencing of BPIFB3 in cells also depleted of beclin-1 enhanced PV replication (Fig. 7D) and PV titers (data not shown). These data suggest that when BPIFB3 is silenced, PV replication can occur in a beclin-1-independent manner. In all experiments, efficacy of silencing was assessed by RT-qPCR and/or immunoblotting (see Fig. S7 in the supplemental material).
DISCUSSION
The physiological function of the BPI fold-containing family has remained elusive. Here we have shown that BPIFB3 is associated with the ER, where its RNAi-mediated silencing enhances basal AP formation but does not promote autophagic flux. In contrast, overexpression of BPIFB3 induces the formation of enlarged LC3B vacuoles and prevents LC3B lipidation in response to rapamycin treatment. Interestingly, we found that knockdown of BPIFB3 greatly altered the morphology of CVB-induced replication organelles and enhanced CVB replication in a manner independent of the core autophagic initiation machinery (beclin-1, ATG14, and UVRAG). These studies point to a previously uncharacterized regulatory role for BPIFB3 in normal cellular autophagy and also suggest a specific role for this protein in the control of autophagy during CVB replication. A model of the pathway by which BPIFB3 controls CVB replication is shown in Fig. 7E.
We found that depletion of BPIFB3 induced AP formation and led to the formation of APs and IMs along the ER membrane. Importantly, these APs and IMs appeared in cells under nutrient-rich conditions and occurred only as a consequence of BPIFB3 depletion. Given that we did not observe any reduction in the levels of p62 in BPIFB3si-transfected cells under nutrient-rich conditions, loss of BPIFB3 likely does not augment flux, but more likely negatively regulates IM formation, whereby its depletion enhances the initiation of autophagy and AP formation, possibly by the destabilization of ER-associated membranes. Consistent with a role for BPIFB3 in the early events associated with autophagy, we found that overexpression of BPIFB3 inhibited rapamycin-induced autophagic signaling and led to the dramatic formation of enlarged vacuoles when coexpressed with LC3B. In addition, overexpression of BPIFB3 inhibited the cleavage and lipidation of LC3B in response to rapamycin treatment. LC3 is cleaved by the ATG4 protease and is then conjugated to phosphatidylethanolamine (PE) by ATG7/ATG3 (to form LC3-II). Only the cleaved and PE-conjugated form of LC3 associates with the AP (on both the inner and outer membranes). Thus, our data suggest that expression of BPIFB3 inhibits an early autophagic event, before the formation of LC3-II. The vacuoles formed in BPIFB3-overexpressing cells may represent enlarged IMs that are unable to mature into APs, thus inhibiting the formation LC3B-II. Enlarged LC3B vacuoles have been observed previously under conditions in which autophagic flux is inhibited (37). However, unlike these reports, we did not observe the accumulation of LC3B-II or p62 in cells overexpressing BPIFB3, supporting an earlier role for BPIFB3 in the autophagy pathway.
Although our data implicate a role for BPIFB3 in early events associated with autophagy, they also suggest that BPIFB3 may facilitate some aspect of AP-lysosome fusion. In support of this, we found that LC3B puncta formed in BPIFB3si-transfected cells were highly associated with both p62 and LAMP2, suggesting that these vesicles are not degradative in nature. Similar to our findings, others have shown that RNAi-mediated silencing of βCOP leads to the accumulation of APs, without any corresponding reduction in the levels of p62 (38). In addition, APs formed in βCOP-depleted cells are not acidic or degradative, despite their colocalization with markers of lysosomes such as LAMP2 (38). These characteristics are strikingly similar to those observed in BPIFB3-depleted cells, in which we observed increased AP numbers without a corresponding enhancement of autophagic flux and an absence of degradative APs. The inhibition of AP maturation in βCOP-depleted cells results from defects in early endosomal pathways and correlates with the inability of APs to fuse with early endosomes, thus altering their maturation (38). Given that depletion of BPIFB3 also exerts significant alterations in early and late endosomes, as well as inducing the formation of enlarged lysosomes, it seems likely that the increases in APs observed in cells transfected with BPIFB3si may also be a direct consequence of alterations in vesicular trafficking that might alter AP maturation.
We found that expression of the first BPI fold of BPIFB3 was sufficient to induce LC3B vacuoles and strongly colocalized with LC3B puncta and that neither the second BPI fold nor expression of BPIFB6 (or its individual BPI folds) induced these vacuoles. The individual BPI folds of BPIFB3 are predicted to strongly associate with lipids, given its structural similarities to BPI and other members of the BPI fold-containing family. The structure of BPIFB3 has not been solved, but the structure of the related BPI has been determined and is likely to be conserved among members of the related PLUNC family. BPI exists in a boomerang shape and contains two hydrophobic lipid-binding pockets in each fold (39). Given the lipid-binding capabilities of BPIFB3 and its boomerang shape, it may function in some aspect of influencing the membrane architecture of the ER, which could profoundly influence the initiation of autophagy. Growing evidence points to a prominent role for the ER in AP biogenesis, although other cellular organelles have also been implicated in this process. In particular, a subdomain of the ER termed the “omegasome” (for its omega shape) primarily serves as the site of AP initiation, and APs mature only once they have fully emerged from this domain (2). ER tubular structures link the omegasome with the IM (31). Interestingly, we also observed extensive enhancement of ER tubules in cells depleted of BPIFB3 (data not shown), which may alter the propensity of IMs to form along the ER and fully mature into APs.
Similar to the previous work of others (18, 19, 28), our work presented here supports a model in which CVB requires components of the core autophagic machinery for its replication. However, our work also suggests that in cells with low levels of BPIFB3, CVB can induce autophagy independently from many components of the core initiation machinery and that this induction might be more efficient than its utilization of the canonical pathway. Recently, CVB release in extracellular vesicles harboring markers of autophagosomes has been suggested to play a role in CVB release independent of cell lysis (40). Although it is possible that BPIFB3 silencing enhances this process by promoting the induction of autophagy, the dramatic enhancement in CVB replication (with ~500- to 1,000-fold enhancement in CVB viral RNA) seems unlikely to result from alterations in viral egress alone. Instead, our data suggest that loss of BPIFB3 directly promotes the enhancement of a noncanonical form of autophagy that occurs independently from the core initiation components, including beclin-1.
The role of AP-lysosome fusion in CVB replication has remained unclear. Although a previous study suggested that CVB inhibits autophagic flux as a means to facilitate its replication (18), this conclusion was largely based on the enhancement of CVB replication in cells transfected with VAMP2 siRNA. Although VAMP2 plays a role in synaptic vesicle fusion (41), it has not been implicated in AP-lysosome fusion, for which the specific SNAREs were only recently identified (9, 10). Furthermore, p62 has been suggested to be directly cleaved by CVB during its replication (42). However, we did not detect cleavage of p62 (or reductions in p62 levels) in control cells infected with CVB, which could be the result of cell type and/or virus strain differences. Our data showed that RNAi-mediated silencing of BPIFB3 enhances CVB replication, which then sensitizes CVB to the effects of BafA1, suggesting that fusion and/or autophagic flux might facilitate viral replication, but this may not occur when CVB utilizes a canonical form of autophagy. However, further studies on the specific role that autophagic flux plays during CVB replication are required.
Our work presented here provides a detailed analysis of the role of the PLUNC family member BPIFB3 in the regulation of autophagy and CVB replication. We do not yet know whether other members of the PLUNC family also function in a similar manner, but given the conservation of lipid-binding sites within this family, it is possible that other members also function to regulate cellular trafficking and/or autophagy. However, the lack of enhancement of CVB replication upon silencing of other members of the PLUNC family suggests that BPIFB3 is specifically involved in the regulation of autophagy in CVB-infected cells. Moreover, we did not find that silencing of other BPIFB family members (e.g., BPIFB2, BPIFB4, BPIFB6) had any effect on autophagy. Taken together, our findings not only represent a significant step toward identifying the function of a member of the PLUNC family in both the regulation of autophagy and in the life cycle of select enteroviruses, but also in the identification of the only known cellular component whose depletion promotes noncanonical autophagy in CVB-infected cells to promote viral replication.
MATERIALS AND METHODS
Cells and viruses.
Unless otherwise stated, all experiments were performed in human brain microvascular endothelial cells (HBMEC) cultured as described previously (43). U2OS and HeLa cells were cultured as described previously (21, 44). Experiments were performed with CVB3-RD or PV, expanded as described elsewhere (21, 43). Experiments measuring productive virus infection were performed with 1 to 3 PFU/cell for ~16 h unless otherwise stated. Plaque assays were performed as described previously (21). CellLight ER-RFP BacMam 2.0 baculovirus was purchased from Invitrogen.
Antibodies and inhibitors.
Rabbit and goat polyclonal and mouse monoclonal antibodies directed against EEA1 (N-19), Flag/OctA (D-8, H-5), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; FL-335), LAMP-2 (H4B4), Rab7 (H-50), V5 (E10), and GFP (FL, B-2) were purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-Flag (M2) and rabbit polyclonal anti-BPIFB3 antibodies were purchased from Sigma. Mouse monoclonal antibodies to p230/Golgin and calnexin were purchased from BD Biosciences. Mouse anti-enterovirus VP1 (Ncl-Entero) was obtained from Novocastra Laboratories. Rabbit anti-LC3B and mouse anti-p62 and anti-MTC02 antibodies were purchased from Abcam. Rabbit anti-ATG7, ATG14, beclin-1, and UVRAG antibodies was purchased from Cell Signaling Technologies. Mouse anti-double-stranded RNA (anti-dsRNA; J2) antibody was provided by Saumendra Sarkar (University of Pittsburgh). Alexa Fluor-conjugated secondary antibodies were purchased from Invitrogen.
Bafilomycin A1 (3 µM) and spautin-1 (10 µM) were purchased from Sigma. Rapamycin (500 nM) was purchased from Calbiochem.
Plasmids, siRNAs, and transfections.
Flag-tagged BPIFB3 and V5-tagged BPIFB6 were generated by amplification of BPIFB3 or BPIFB6 cDNAs with primers encoding a C-terminal Flag tag (for BPIFB3) and cloned into pcDNA3.1/V5-His TOPO TA per the manufacturer’s instructions (Invitrogen). Virally encoded 3A from CVB or PV was amplified from infected cells and cloned into pcDNA3.1/NT-GFP-TOPO per the manufacturer’s instructions (Invitrogen). BPIFB3 BPI fold 1 (BPI-1) and BPI-2 were amplified from cDNA and cloned into pcDNA3.1/NT-GFP-TOPO per the manufacturer’s instructions (Invitrogen). Primer sequences are available upon request. mRFP-LC3B has been described previously (45). EGFP-LC3B was generated by amplification of LC3B and subsequent cloning into pcDNA3.1/EGFP TOPO TA per the manufacturer’s instructions (Invitrogen). C-terminal EYFP-fused BPIFB3 was constructed by PCR amplification and subsequent cloning into the BamHI and KpnI sites of pEYFP-N1 (Clontech).
Three siRNAs were used to target BPIFB3, BPIFB3-1 (GCUUAACGUGGCCCUGGAUtt), BPIFB3-2 (CCAAAGUGGGCAUGCAUUGtt), and BPIFB3-3 (CCAGUACAUAGAACUGGACtt), and were purchased from Sigma. siRNAs targeting other BPI domain-containing family members, BPIFB2 (CCUGAAAUUCAUUGCUGGUtt), BPIFB4 (CGGGAAGAGUCUUAUUGGCtt), and BPIFB6 (GGAUACAAUGAUUGGUGAGtt), and one targeting ATG7 (GGAGUACAGCUCUUCCUUtt) were purchased from Sigma. Beclin-1, ATG14, and UVRAG siRNAs were purchased from Cell Signaling Technologies. Control (scrambled) siRNAs were purchased from Ambion or Sigma.
Transfections were performed as described previously (45, 46).
Immunofluorescence and electron microscopy.
Cells cultured in chamber slides (Lab-Tek, Nunc) were washed and fixed with either 4% paraformaldehyde (PFA) or with ice-cold methanol. Cells were then permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) and incubated with the indicated primary antibodies for 1 h at room temperature. Following washing, cells were incubated with secondary antibodies for 30 min at room temperature, washed, and mounted with Vectashield (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole (DAPI). The technique utilized for digitonin/Triton X-100 staining was performed as described elsewhere (26). Images were captured using an FV1000 confocal laser scanning microscope (Olympus), analyzed using Imaris (Bitplane), and contrasted and merged using Photoshop (Adobe). Electron microscopy was performed as described previously (45). For calculations of CVB replication organelle size, >100 individual organelles were measured using Imaris for >20 unique cells.
Live-cell imaging.
U2OS cells transfected with the indicated plasmids were plated into glass-bottom 35-mm dishes (Mat-Tek). Approximately 48 h following transfection, plates were placed in a 37°C, CO2-controlled incubator positioned over a motorized inverted microscope to allow for long-term time-lapse imaging (VivaView FL; Olympus), and images were captured every 10 to 15 min for 4 to 8 h.
Immunoblotting.
Cell lysates were prepared with RIPA buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethanesulfonyl fluoride, 1 mg/ml aprotinin, leupeptin, and pepstatin, and 1 mM sodium orthovanadate). Lysates (30 µg) were loaded onto 4-to-20% or 10-to-20% (for LC3B immunoblots) Tris-HCl gels (Bio-Rad, Hercules, CA) and transferred to nitrocellulose or polyvinylidene difluoride membranes. Membranes were blocked in 5% nonfat dry milk, probed with the indicated antibodies, and developed with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) and SuperSignal West Pico or West Dura chemiluminescent substrates (Pierce Biotechnology). Densitometry was performed using ImageJ.
Some immunoblot assays were conducted using an Odyssey infrared imaging system (LI-COR Biosciences). In these cases, membranes were incubated in 5% nonfat dry milk and then incubated with the appropriate antibodies overnight at 4°C. Following washing, membranes were incubated with anti-rabbit or anti-mouse antibodies conjugated to IRDye 680LT or 800CW in the presence of 0.01% SDS and visualized with the Odyssey infrared imaging system. Densitometry was performed using Image Studio.
Subcellular fractionation.
Stable U2OS cells expressing BPIFB3-Flag were grown in a T-150 flask and detached by incubation in homogenization buffer (0.25 M sucrose, 1 mM EDTA, 20 mM HEPES-NaOH; pH 7.4) for 30 min at 4°C and then disrupted by passage through a 22-gauge needle. Homogenates were centrifuged at 3,000 × g for 10 min, and supernatants were collected and centrifuged at 17,000 × g for 15 min. Pellets were resuspended in homogenization buffer adjusted to 35% (wt/vol) Opti-prep and homogenized with a Dounce homogenizer. Homogenates were overlaid onto a 10-to-30% continuous iodixanol gradient and centrifuged at 100,000 × g in a Beckman Coulter microultracentrifuge swinging bucket SW60Ti rotor for 60 min. Sequential 250-µl fractions were then collected from the top of the gradient, and proteins were resolved by immunoblotting.
RT-qPCR.
For cellular mRNA analysis, total RNA was extracted using TRI reagent (MRC, Cincinnati, OH) or a GenElute total RNA miniprep kit (Sigma) according to the manufacturer’s protocol. RNA samples were treated with RNase-free DNase (Qiagen or Sigma). Total RNA was reverse transcribed by using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). For each sample, 1 µg RNA was used for cDNA synthesis. RT-qPCR was performed using iQ SYBR green supermix (Bio-Rad) in an Applied Biosystems StepOnePlus real-time PCR machine. Gene expression was calculated using the 2−ΔΔCT method (47), normalized to human beta-actin. QuantiTect primers against BPIFB2, BPIFB3, BPIFB4, and BPIFB6 were purchased from Qiagen. Primer sequences for ATG7, UVRAG, and PV were as follows: ATG7 (5′-AGATTGTCCTAAAGCAGTTG-3′ and 5′-CCATACATTCACTGAGGTTC-3′); UVRAG (5′-ATGCCAGACCGTCTTGATACA-3′ and 5′-TGACCCAAGTATTTCAGCCCA-3′); PV (5′-CCCCTGAAtgCGGCTAATC-3′ and 5′-GATTGTCACCATAAGCAGC-3′). Actin, beclin-1, and CVB primer sequences have been described elsewhere (45, 46).
Statistical analysis.
Data are presented as means ± standard deviations unless otherwise stated and were analyzed with Prism software (GraphPad) by using the two-tailed unpaired Student’s t test. Experiments were performed a minimum of three times.
SUPPLEMENTAL MATERIAL
Real-time imaging of U2OS cells transfected with BPIFB3-EYFP and mRFP-LC3B. Approximately 48 h posttransfection, cells were treated with rapamycin for ~2 h and images were captured every 10 min for ~5 h. Shown is a movie of mRFP-LC3B puncta. Download
Real-time imaging of U2OS cells transfected with vector (pcDNA3.1) and mRFP-LC3B. At approximately 48 h posttransfection, cells were treated with rapamycin for ~2 h and images were captured every 10 min for ~5 h. Shown is a movie of mRFP-LC3B puncta. Download
CVB infection (black bars) in HBMEC transfected with three independent BPIFB3 siRNAs (BPIFB3si-1, -2, and -3) individually, in combination (1 + 2 + 3), or in cells transfected with control siRNA (CONsi), assessed by immunofluorescence microscopy. Gray bars, BPIFB3 expression, assessed by RT-qPCR. Download
(A) U2OS cells transfected with Flag-BPIFB3 were fixed at ~48 h posttransfection and immunostained with anti-early endosome antigen-1 (EEA1; to label early endosomes; red) (top left) or incubated with Bodipy 493/503 to label lipid droplets (green; top right) and anti-Flag (green or red, as indicated). (B) Subcellular fractionation of cells stably expressing BPIFB3-Flag. Cystosolic, membrane/organelles, nuclear, and cytoskeletal fractions were isolated and probed with antibodies against Flag (BPIFB3, top), calnexin (CXN), cadherins (CAD), c-JUN, and GAPDH. (C) Wild-type or mutant AAEL BPIFB3-Flag in U2OS cells was transiently expressed in U2OS cells, and at ~24 h posttransfection, cells were infected with ER-RFP baculovirus for 24 h. Cells were then immunostained for Flag (green). Download
(A and B) HeLa (A) or 786-O (B) cells transfected with control (CONsi) or BPIFB3 (BPIFB3si) siRNAs for ~48 h were immunostained for LC3B (green). (C) Quantification of the number of LC3B punctae per cell in HeLa or 786-O cells transfected with CONsi or BPIFB3si. A total of ~50 cells were quantified. Download
(A and B) Quantification of the size (A) and numbers (B) of EEA1-, LAMP2 -, and Rab7-positive vesicles in cells transfected with CONsi (black bars) or BPIFB3si (gray bars). Data are shown as mean ± standard deviation. *, P < 0.001. (C) HeLa cells transfected with CONsi or BPIFB3si were fixed and stained with antibodies against LAMP2 (green) and EEA1 (red) at ~48 h posttransfection. Download
(A) Quantification of the percentage of cells displaying enlarged vacuoles in cells transfected with either vector (black bars) or BPIFB3-Flag (gray bars) and EGFP-LC3B, mRFP-LC3B, or mRFP-LAMP1 under nutrient-rich conditions. Data are shown as mean ± standard deviation. (B) U2OS cells transfected with BPIFB6-V6 and mRFP-LC3B were fixed and immunostained for V5 (in green) at ~48 h posttransfection. (C) U2OS cells transfected with vector or BPIFB3-Flag and mRFP-LAMP1 were fixed and immunostained for Flag (in green) at ~48 h posttransfection. Download
(A) Select frames (taken at 10-min intervals) from time-lapse live-cell imaging of U2OS cells transfected with vector and mRFP-LC3B and treated with rapamycin from ~60 min posttreatment. See Movie S2 here in the supplemental material for the complete movie. (B) U2OS cells transfected with EGFP-BPI-1 and mRFP-LC3B for ~48 h were fixed. Download
(A) Immunoblots for ATG7 (top left), ATG14 (top right), beclin-1 (bottom left), and UVRAG (bottom right) in HBMEC transfected with CONsi or ATG7si, ATG14si, BECLN1si, or UVRAGsi, as indicated. At the bottom of all panels, GAPDH immunoblots are shown as loading controls. (B) RT-qPCR for ATG7, BECLN1, or UVRAG in HBMEC transfected with CONsi or BPIFB3si and either ATG7si, BECLN1si, or UVRAGsi, as indicated. Data are shown as mean ± standard deviation. *, P < 0.05. Download
ACKNOWLEDGMENTS
We thank Kwang Sik Kim (Johns Hopkins University) for human brain microvascular endothelial cells, Saumendra Sarkar for dsRNA antibody, Ming Sun, Jana Jacobs, and Coyne Drummond (all from the University of Pittsburgh) for technical assistance, and Jeffrey Bergelson (Children’s Hospital of Philadelphia) for careful review of the manuscript.
This project was supported by the NIH R01-AI081759 (C.B.C.). In addition, C.B.C. is a recipient of the Burroughs Wellcome Investigators in the pathogenesis of Infectious Disease Award.
Footnotes
Citation Delorme-Axford E, Morosky S, Bomberger J, Stolz DB, Jackson WT, Coyne CB. 2014. BPIFB3 regulates autophagy and coxsackievirus B replication through a noncanonical pathway independent of the core initiation machinery. mBio 5(6):e02147-14. doi:10.1128/mBio.02147-14.
REFERENCES
- 1. Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437. 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 2. Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van der Vaart A, Griffith J, Reggiori F. 2010. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol. Biol. Cell 21:2270–2284. 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ge L, Melville D, Zhang M, Schekman R. 2013. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife 2:e00947. 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141:656–667. 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature 495:389–393. 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 7. Feng Y, He D, Yao Z, Klionsky DJ. 2014. The machinery of macroautophagy. Cell Res. 24:24–41. 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parzych KR, Klionsky DJ. 2014. An overview of autophagy: morphology, mechanism, and regulation. Antioxid. Redox Signal. 20:460–473. 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furuta N, Fujita N, Noda T, Yoshimori T, Amano A. 2010. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol. Biol. Cell 21:1001–1010. 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Itakura E, Kishi-Itakura C, Mizushima N. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151:1256–1269. 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 11. Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. 2008. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 15:1318–1329. 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 12. Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. 2007. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am. J. Pathol. 170:75–86. 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. 2009. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461:654–658. 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 14. Schuck S, Gallagher CM, Walter P. 2014. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 127:4078–4088. 10.1242/jcs.154716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schlegel A, Giddings TH, Jr, Ladinsky MS, Kirkegaard K. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suhy DA, Giddings TH, Jr, Kirkegaard K. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953–8965. 10.1128/JVI.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLOS Biol. 3:e156. 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 82:9143–9153. 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alirezaei M, Flynn CT, Wood MR, Whitton JL. 2012. Pancreatic acinar cell-specific autophagy disruption reduces coxsackievirus replication and pathogenesis in vivo. Cell Host Microbe 11:298–305. 10.1016/j.chom.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richards AL, Jackson WT. 2012. Intracellular vesicle acidification promotes maturation of infectious poliovirus particles. PLoS Pathog. 8:e1003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coyne CB, Bozym R, Morosky SA, Hanna SL, Mukherjee A, Tudor M, Kim KS, Cherry S. 2011. Comparative RNAi screening reveals host factors involved in enterovirus infection of polarized endothelial monolayers. Cell Host Microbe 9:70–82. 10.1016/j.chom.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bingle CD, Bingle L, Craven CJ. 2011. Distant cousins: genomic and sequence diversity within the BPI fold-containing (BPIF)/plunc protein family. Biochem. Soc. Trans. 39:961–965. 10.1042/BST0390961. [DOI] [PubMed] [Google Scholar]
- 23. Andrault JB, Gaillard I, Giorgi D, Rouquier S. 2003. Expansion of the BPI family by duplication on human chromosome 20: characterization of the RY gene cluster in 20q11.21 encoding olfactory transporters/antimicrobial-like peptides. Genomics 82:172–184. 10.1016/S0888-7543(03)00102-2. [DOI] [PubMed] [Google Scholar]
- 24. Dear TN, Boehm T, Keverne EB, Rabbitts TH. 1991. Novel genes for potential ligand-binding proteins in subregions of the olfactory mucosa. EMBO J. 10:2813–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raykhel I, Alanen H, Salo K, Jurvansuu J, Nguyen VD, Latva-Ranta M, Ruddock L. 2007. A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol. 179:1193–1204. 10.1083/jcb.200705180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saito K, Chen M, Bard F, Chen S, Zhou H, Woodley D, Polischuk R, Schekman R, Malhotra V. 2009. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell 136:891–902. 10.1016/j.cell.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 27. Delorme-Axford E, Sadovsky Y, Coyne CB. 2013. Lipid raft- and SRC family kinase-dependent entry of coxsackievirus B into human placental trophoblasts. J. Virol. 87:8569–8581. 10.1128/JVI.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, Whitton JL. 2010. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J. Virol. 84:12110–12124. 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614. 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 31. Uemura T, Yamamoto M, Kametaka A, Sou YS, Yabashi A, Yamada A, Annoh H, Kametaka S, Komatsu M, Waguri S. 2014. A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol. Cell. Biol. 34:1695–1706. 10.1128/MCB.01327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dunn WA., Jr. 1990. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J. Cell Biol. 110:1923–1933. 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. 2008. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 4:849–850. 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 34. Saftig P, Beertsen W, Eskelinen EL. 2008. LAMP-2: a control step for phagosome and autophagosome maturation. Autophagy 4:510–512. 10.4161/auto.5724. [DOI] [PubMed] [Google Scholar]
- 35. Bayer N, Schober D, Prchla E, Murphy RF, Blaas D, Fuchs R. 1998. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. J. Virol. 72:9645–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. 1991. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 266:17707–17712. [PubMed] [Google Scholar]
- 37. Choi YJ, Park YJ, Park JY, Jeong HO, Kim DH, Ha YM, Kim JM, Song YM, Heo HS, Yu BP, Chun P, Moon HR, Chung HY. 2012. Inhibitory effect of mTOR activator MHY1485 on autophagy: suppression of lysosomal fusion. PLoS One 7:e43418. 10.1371/journal.pone.0043418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Razi M, Chan EY, Tooze SA. 2009. Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 185:305–321. 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beamer LJ, Carroll SF, Eisenberg D. 1997. Crystal structure of human BPI and two bound phospholipids at 2.4-angstrom resolution. Science 276:1861–1864. 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 40. Robinson SM, Tsueng G, Sin J, Mangale V, Rahawi S, McIntyre LL, Williams W, Kha N, Cruz C, Hancock BM, Nguyen DP, Sayen MR, Hilton BJ, Doran KS, Segall AM, Wolkowicz R, Cornell CT, Whitton JL, Gottlieb RA, Feuer R. 2014. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 10:e1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schoch S, Deák F, Königstorfer A, Mozhayeva M, Sara Y, Südhof TC, Kavalali ET. 2001. SNARE function analyzed in synaptobrevin/VAMP knockout mice. Science 294:1117–1122. 10.1126/science.1064335. [DOI] [PubMed] [Google Scholar]
- 42. Shi J, Wong J, Piesik P, Fung G, Zhang J, Jagdeo J, Li X, Jan E, Luo H. 2013. Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFkB signaling. Autophagy 9:1591–1603. 10.4161/auto.26059. [DOI] [PubMed] [Google Scholar]
- 43. Coyne CB, Kim KS, Bergelson JM. 2007. Poliovirus entry into human brain microvascular cells requires receptor-induced activation of SHP-2. EMBO J. 26:4016–4028. 10.1038/sj.emboj.7601831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bozym RA, Delorme-Axford E, Harris K, Morosky S, Ikizler M, Dermody TS, Sarkar SN, Coyne CB. 2012. Focal adhesion kinase is a component of antiviral RIG-I-like receptor signaling. Cell Host Microbe 11:153–166. 10.1016/j.chom.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Delorme-Axford E, Donker RB, Mouillet JF, Chu T, Bayer A, Ouyang Y, Wang T, Stolz DB, Sarkar SN, Morelli AE, Sadovsky Y, Coyne CB. 2013. Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl. Acad. Sci. U. S. A. 110:12048–12053. 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jacobs JL, Zhu J, Sarkar SN, Coyne CB. 2014. Regulation of mitochondrial antiviral signaling (MAVS) expression and signaling by the mitochondria-associated endoplasmic reticulum membrane (MAM) protein Gp78. J. Biol. Chem. 289:1604–1616. 10.1074/jbc.M113.520254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time imaging of U2OS cells transfected with BPIFB3-EYFP and mRFP-LC3B. Approximately 48 h posttransfection, cells were treated with rapamycin for ~2 h and images were captured every 10 min for ~5 h. Shown is a movie of mRFP-LC3B puncta. Download
Real-time imaging of U2OS cells transfected with vector (pcDNA3.1) and mRFP-LC3B. At approximately 48 h posttransfection, cells were treated with rapamycin for ~2 h and images were captured every 10 min for ~5 h. Shown is a movie of mRFP-LC3B puncta. Download
CVB infection (black bars) in HBMEC transfected with three independent BPIFB3 siRNAs (BPIFB3si-1, -2, and -3) individually, in combination (1 + 2 + 3), or in cells transfected with control siRNA (CONsi), assessed by immunofluorescence microscopy. Gray bars, BPIFB3 expression, assessed by RT-qPCR. Download
(A) U2OS cells transfected with Flag-BPIFB3 were fixed at ~48 h posttransfection and immunostained with anti-early endosome antigen-1 (EEA1; to label early endosomes; red) (top left) or incubated with Bodipy 493/503 to label lipid droplets (green; top right) and anti-Flag (green or red, as indicated). (B) Subcellular fractionation of cells stably expressing BPIFB3-Flag. Cystosolic, membrane/organelles, nuclear, and cytoskeletal fractions were isolated and probed with antibodies against Flag (BPIFB3, top), calnexin (CXN), cadherins (CAD), c-JUN, and GAPDH. (C) Wild-type or mutant AAEL BPIFB3-Flag in U2OS cells was transiently expressed in U2OS cells, and at ~24 h posttransfection, cells were infected with ER-RFP baculovirus for 24 h. Cells were then immunostained for Flag (green). Download
(A and B) HeLa (A) or 786-O (B) cells transfected with control (CONsi) or BPIFB3 (BPIFB3si) siRNAs for ~48 h were immunostained for LC3B (green). (C) Quantification of the number of LC3B punctae per cell in HeLa or 786-O cells transfected with CONsi or BPIFB3si. A total of ~50 cells were quantified. Download
(A and B) Quantification of the size (A) and numbers (B) of EEA1-, LAMP2 -, and Rab7-positive vesicles in cells transfected with CONsi (black bars) or BPIFB3si (gray bars). Data are shown as mean ± standard deviation. *, P < 0.001. (C) HeLa cells transfected with CONsi or BPIFB3si were fixed and stained with antibodies against LAMP2 (green) and EEA1 (red) at ~48 h posttransfection. Download
(A) Quantification of the percentage of cells displaying enlarged vacuoles in cells transfected with either vector (black bars) or BPIFB3-Flag (gray bars) and EGFP-LC3B, mRFP-LC3B, or mRFP-LAMP1 under nutrient-rich conditions. Data are shown as mean ± standard deviation. (B) U2OS cells transfected with BPIFB6-V6 and mRFP-LC3B were fixed and immunostained for V5 (in green) at ~48 h posttransfection. (C) U2OS cells transfected with vector or BPIFB3-Flag and mRFP-LAMP1 were fixed and immunostained for Flag (in green) at ~48 h posttransfection. Download
(A) Select frames (taken at 10-min intervals) from time-lapse live-cell imaging of U2OS cells transfected with vector and mRFP-LC3B and treated with rapamycin from ~60 min posttreatment. See Movie S2 here in the supplemental material for the complete movie. (B) U2OS cells transfected with EGFP-BPI-1 and mRFP-LC3B for ~48 h were fixed. Download
(A) Immunoblots for ATG7 (top left), ATG14 (top right), beclin-1 (bottom left), and UVRAG (bottom right) in HBMEC transfected with CONsi or ATG7si, ATG14si, BECLN1si, or UVRAGsi, as indicated. At the bottom of all panels, GAPDH immunoblots are shown as loading controls. (B) RT-qPCR for ATG7, BECLN1, or UVRAG in HBMEC transfected with CONsi or BPIFB3si and either ATG7si, BECLN1si, or UVRAGsi, as indicated. Data are shown as mean ± standard deviation. *, P < 0.05. Download