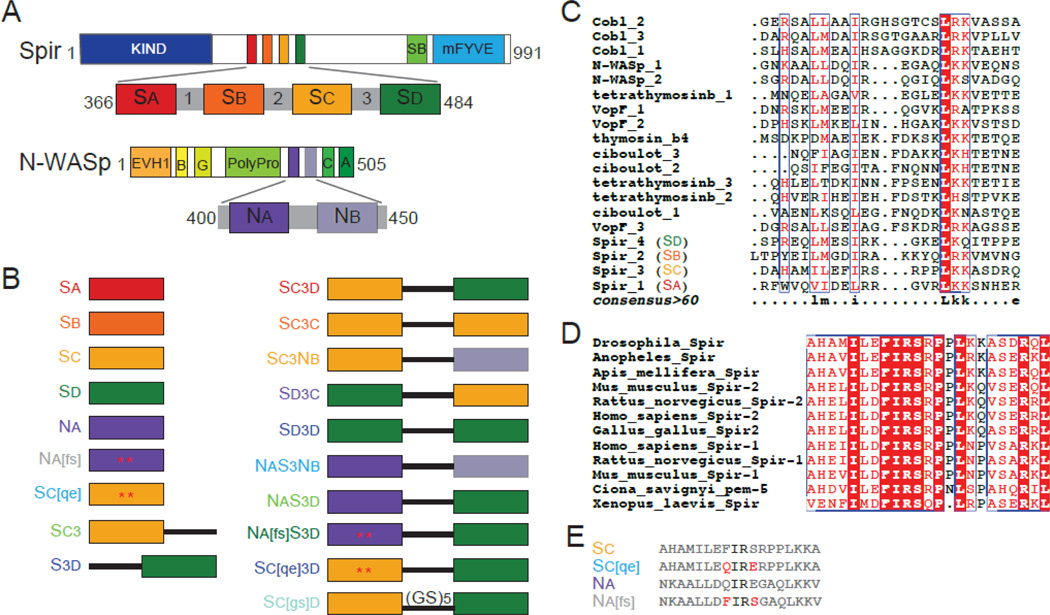
Figure 1.
Spir and N-WASp domains and constructs used. (A) Schematic of Drosophila Spir domain structure: KIND, kinase noncatalytic C-lobe domain (dark blue), , Spir box (light green), mFYVE, modified Fab1/YOTB/Vac1/EEA1 zinc-binding domain (light blue). Expanded: WH2, WASp homology-2 cluster (red, orange, yellow, and dark green) and linker domain regions (grey). Schematic of Rattus N-WASp domain structure: EVH1, Ena/VASP homology-1 domains (orange), B, basic region (light orange), G, GTPase binding domain (yellow), PolyPro, proline rich region (light green), C, central region (green), A, acidic domain (dark green). Expanded: WH2 domains (dark and light purple). (B) Schematic of the constructs used. Colored boxes represent the WH2 domains reported in (A), the black line represents Linker 3 from Spir, and mutated residues are indicated by an asterisk and shown is (E). The color of the construct name is used in future figures. (C) We compared the amino acid sequence of WH2 domains from proteins containing tandem WH2 domains with ClustalW [44]. The bottom line (consensus>60) reflects the consensus as follows: uppercase is identity; lowercase is consensus level great than 60%. (D) Alignment of Sc from nine species of Spir. The entire domain is highly conserved, including the residues flanking the isoleucine and arginine commonly found in the alpha helix of WH2 domains. (E) Sequences of the mutated WH2 domains and their wild type counterparts. Mutated residues in red are represented by asterisks in (B).