Abstract
An effective vaccine against human immunodeficiency virus type 1 (HIV-1) will have to provide protection against a vast array of different HIV-1 strains. Current methods to measure HIV-1-specific binding antibodies following immunization typically focus on determining the magnitude of antibody responses, but the epitope diversity of antibody responses has remained largely unexplored. Here we describe the development of a global HIV-1 peptide microarray that contains 6,564 peptides from across the HIV-1 proteome and covers the majority of HIV-1 sequences in the Los Alamos National Laboratory global HIV-1 sequence database. Using this microarray, we quantified the magnitude, breadth, and depth of IgG binding to linear HIV-1 sequences in HIV-1-infected humans and HIV-1-vaccinated humans, rhesus monkeys and guinea pigs. The microarray measured potentially important differences in antibody epitope diversity, particularly regarding the depth of epitope variants recognized at each binding site. Our data suggest that the global HIV-1 peptide microarray may be a useful tool for both preclinical and clinical HIV-1 research.
Keywords: HIV, peptide microarray, diversity, antibody, vaccine
1. Introduction
One of the fundamental challenges in HIV-1 vaccine development is the tremendous diversity of HIV-1 strains worldwide (Korber et al., 2001; Gaschen et al., 2002; Taylor et al., 2008; Barouch and Korber, 2009; Korber et al., 2009; Walker et al., 2011; Ndung'u and Weiss, 2012; Picker et al., 2012; Stephenson and Barouch, 2013). Globally, there are more than a dozen HIV-1 subtypes and hundreds of circulating HIV-1 recombinant forms (CRFs), and between-subtype variation can be as large as 35% (Hemelaar et al., 2006; Taylor et al., 2008; Ndung’u and Weiss, 2012). Several HIV-1 vaccine candidates under development aim to overcome the challenge of HIV-1 genetic diversity either through the choice of HIV-1 antigen sequence or the method of antigen delivery (Stephenson and Barouch, 2013). However, most tools used to assess the immunogenicity of these vaccines focus on measuring the magnitude of HIV-1-specific antibody responses, rather than the epitope diversity and specificity of these responses.
Peptide microarrays are a potential tool for the measurement of antibody diversity against linear epitopes in HIV-1 vaccine studies. This platform has been utilized to characterize antibody binding to linear sequences in multiple fields, including HIV-1 vaccine research (Nahtman et al., 2007; Cerecedo et al., 2008; Gaseitsiwe et al., 2008; Lorenz et al., 2009; Tomaras et al., 2011; Haynes et al., 2012). HIV-1-specific microarrays to date, however, have not included extensive coverage of variable sequences (Karasavvas et al., 2012; Gottardo et al., 2013; Imholte et al., 2013). Here we describe the development of a global HIV-1 peptide microarray that includes 6,564 overlapping linear HIV-1 peptides covering most common HIV-1 variants in the HIV-1 sequence database at Los Alamos National Laboratory (LANL). This microarray includes 6,564 peptides, including an average of 7 peptide variants for each 15 amino acid position in HIV-1 Env, Gag, Nef, Pol, Rev, Tat, and Vif, with up to 95 peptide variants per location within the most variable regions of HIV-1 Env. This epitope diversity on the microarray allows for more precise measurements of the magnitude, breadth and depth of HIV-1-specific binding IgG responses.
2. Methods
2.1.Generation of peptide library
In collaboration with JPT Peptide Technologies (Berlin, Germany), we designed a library of HIV-1 linear peptides that provided optimal coverage of HIV-1 global sequence diversity. We began by downloading the sequence alignment for HIV-1 genes ENV, GAG, NEF, POL, REV, TAT, and VIF from the website of the LANL HIV-1 sequence database (Theoretical Biology and Biophysics, 2009) using the following settings: Alignment type: Web Alignment (all complete sequences); Year: 2009; Region: Pre-defined region of the genome; Subtype: All M Group (A-K + Recombinants); DNA/Protein: Protein; Format: FASTA. Full length proteins of gp120, gp41, p17, p24, Tat, and Nef were used, as were the immunogenic fragments of p2p7p1p6, protease, reverse transcriptase, integrase, Vif, and Ref as published by LANL (Theoretical Biology and Biophysics, 2010) (Table 1).
Table 1.
Composition of global HIV-1 peptide microarray.
| GENE | Position HXB2 Start |
Position HXB2 End |
Protein | # of Input Sequences |
# Peptides on Array |
Global Coverage [%] |
|---|---|---|---|---|---|---|
| ENV | 1 | 511 | gp160_1 | 2248 | 2672 | 50.2 |
| 512 | 856 | gp160_2 | 2248 | 1210 | 65.5 | |
| GAG | 1 | 132 | p17 | 3578 | 517 | 58.4 |
| 133 | 363 | p24 | 3578 | 264 | 86.2 | |
| 364 | 379 | p2p7p1p6_1 | 3578 | 20 | 10.5 | |
| 380 | 394 | p2p7p1p6_2 | 3578 | 18 | 17.9 | |
| 406 | 420 | p2p7p1p6_3 | 3578 | 13 | 71.7 | |
| 428 | 450 | p2p7p1p6_4 | 3577 | 50 | 68.7 | |
| NEF | 1 | 205 | Nef | 2606 | 667 | 55.3 |
| POL | 57 | 71 | Prot_1 | 1765 | 10 | 88.4 |
| 97 | 104 | Prot_2 | 1765 | 13 | 75.4 | |
| 217 | 231 | RT_1 | 1765 | 4 | 90.5 | |
| 402 | 420 | RT_2 | 1765 | 8 | 69.3 | |
| 447 | 461 | RT_3 | 1765 | 13 | 51.4 | |
| 500 | 514 | RT_4 | 1765 | 14 | 86.1 | |
| 529 | 543 | RT_5 | 1765 | 22 | 84.9 | |
| 594 | 608 | RT_6 | 1765 | 17 | 52 | |
| 673 | 707 | RT_7 | 1765 | 51 | 41.5 | |
| 716 | 774 | Int_1 | 1765 | 98 | 40 | |
| 865 | 879 | Int_2 | 1765 | 16 | 70.3 | |
| 924 | 990 | Int_3 | 1765 | 68 | 81.3 | |
| REV | 3 | 17 | Rev_1 | 1370 | 17 | 26.1 |
| 32 | 50 | Rev_2 | 1370 | 14 | 78.3 | |
| 70 | 116 | Rev_3 | 1370 | 187 | 42.6 | |
| TAT | 1 | 100 | Tat | 1286 | 296 | 55.5 |
| VIF | 33 | 47 | Vif_1 | 1643 | 19 | 9.8 |
| 174 | 192 | Vif_2 | 1642 | 38 | 38.7 | |
From the global sequence database, we selected the individual sequences to be used as peptides that would provide optimal coverage of sequence diversity using the program package MosaicVaccines.1.2.11 from LANL (ftp://ftp-t10.lanl.gov/pub/btk/mosaic/) (Fischer et al., 2007a; Thurmond et al., 2008a). Parameters for the generation of MOSAIC sequences were –s 20 –d=true –T 20 –p 100. Sequence manipulation and processing were performed in R 2.11.1 (http://www.r-project.org/) using the package Biostrings (http:www.bioconductor.org/packages/2.2/bioc/html/Biostrings.html) or using bespoke scripts in python (http:www.python.org/).
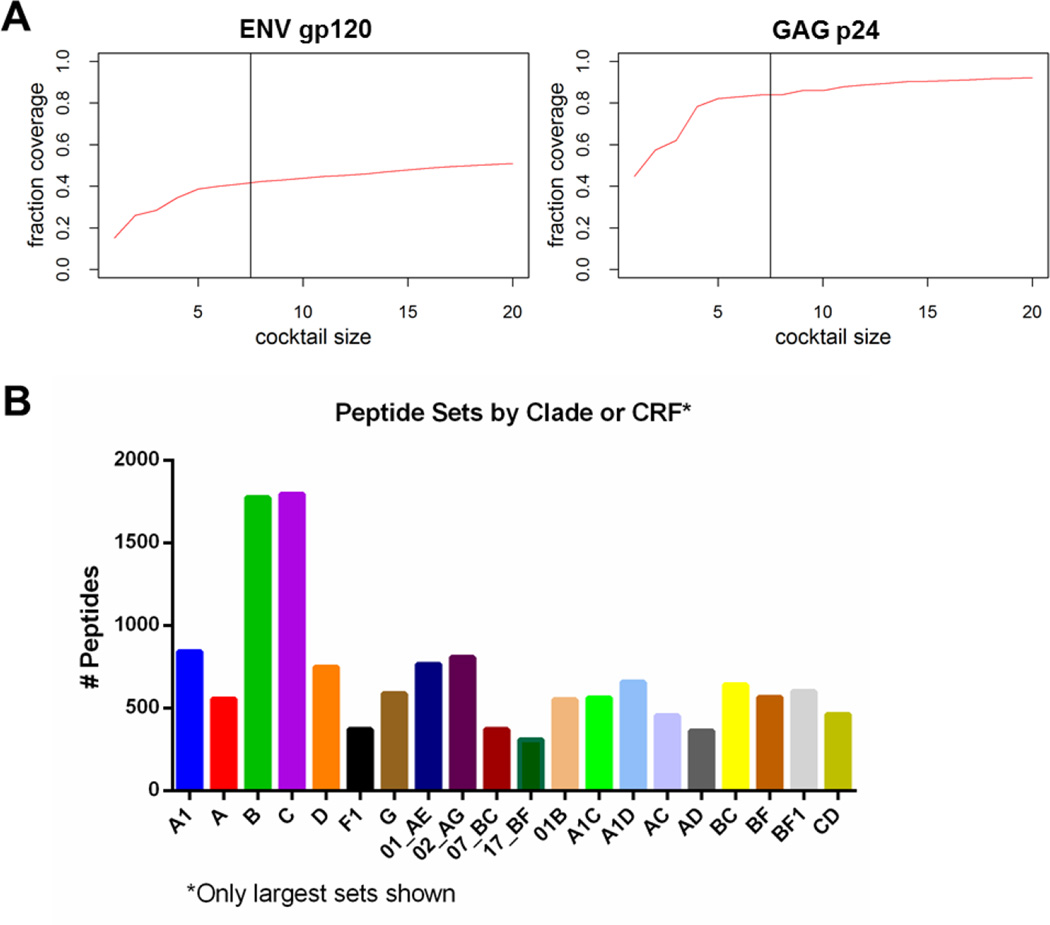
Since our goal was to cover the seven most frequent clades (A, B, C, D, G, CRF01_AE, and CRF02_AG), we used a stepwise approach to generating an optimal sequence cocktail. As a first step, the MOSAIC program was used to identify a sequence for each gene product or fragment from each of the 7 most frequent clades, and the resulting 7 sequences were merged into one cocktail. Secondly, we identified 13 additional sequences which showed best coverage without consideration of the clade. These two sequence cocktails were merged into one cocktail and evaluated for gain of coverage for each sequence. All sequences which did not gain more than 0.75% of coverage were removed from the cocktail. Thirdly, MOSAIC sequences were generated for each gene product or fragment, respectively (Fischer et al., 2007b; Thurmond et al., 2008b). For the MOSAIC runs the sequence cocktails generated in the previous step were used as fixed sequences. The resulting cocktails were evaluated in terms of coverage gain. All MOSAIC cocktails which gained less than 1% coverage were removed, and a maximum of 2 MOSAIC sequences was kept in the final cocktail. Figure 1A displays the relationship between the increasing size of the cocktail and the plateauing increase in coverage for gp120.
Figure 1. Generation of a global HIV-1 peptide library.
(A) Cocktail size of Env gp120 and Gag p24 sequences is plotted against percent coverage of global sequence variability. The cocktails consist of the single best covering sequences from each of the 7 frequent clades (left of vertical line), and the 13 best covering sequences as calculated from the complete HIV-1 sequence library and MOSAIC sequences (right of vertical line). (B) Clade- and circulating recombinant form (CRF)-specific peptide sets are shown. Only sets that have >300 peptides are shown out of a total of 135 sets. If a peptide sequence was found in multiple clades/CRFs, then it was counted in multiple sets.
Once we had generated a cocktail of sequences with optimal global coverage, we then generated a library of peptides where all sequences within the cocktail were covered at a minimal number of peptides. One of the sequences was used as a template sequence and processed into 15 amino acid peptides overlapping by 11 amino acids. All other sequences within the cocktail were fragmented into peptide scans of 15 amino acid peptides overlapping by 14 amino acids. Of note, this length of peptide (15 amino acids) covers 83% of known linear antibody epitopes in the LANL immunology database, including the median length of epitopes (11 amino acids) (Theoretical Biology and Biophysics, 2014). Scan-peptides were then aligned onto the scan-peptides of the template. The resulting 5,141 peptides covered all template sequences completely. For ENV, we performed one additional step to assure that every region of the protein was represented on the microarray by adding additional MOSAIC sequences that our group generated in the course of HIV-1 vaccine design (Barouch et al., 2010; Barouch et al., 2013). To overcome the bias of peptides towards conserved regions of the protein, we also included an additional 1,004 peptides from the variable loops V2 and V3 of gp120 in the library.
The final library consisted of 6,654 peptides from 135 different clades or CRFs. CRFs are circulating related variants that have different regions associated with the different major HIV-1 clades (Robertson et al., 2000); for example CRF_02 has sections of clade A and clade G, and is common in West Africa. (For a comprehensive listing of CRF’s see http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html). By capturing A clade diversity, we capture some of the diversity found in the regions of CRF_02 that are A-like, but CRF_02 started with a recombinant founder virus decades ago, and has been spreading and diversifying as a separate lineage (Zhang et al., 2010), and so it will have its own distinctive evolutionary trajectory. Each CRF represents its own lineage, thus by including the CRFs in diversity considerations, not just major clades, we take a more comprehensive and realistic view of global diversity than by a more narrow examination of major clades. Figure 1B shows how many peptides were included in each clade- or CRF-specific peptide set (only sets that contain >300 peptides are shown). If a peptide sequence was found in multiple clades/CRFs, then it was counted in multiple sets. Peptides sets from the seven most frequent clades (A, B, C, D, G, CRF_01, and CRF_02) include >500 peptides each.
2.2 Microarray printing
PepStar peptide microarrays were produced by JPT Peptide Technologies GmbH (Berlin, Germany). All peptides were synthesized on cellulose membranes using SPOT synthesis technology. Subsequent to a final synthesis step attaching a reactivity tag to each peptide’s N-terminus, the side chains were deprotected and the solid-phase bound peptides were transferred into 96-well microtiter filtration plates (Millipore, Bedford, MA, USA). For cleaving the peptides from the cellulose membrane the individual spots were treated with aqueous triethylamine [2.5% (v/v)]. The peptide-containing solution was centrifuge-filtered into daughter plates and the solvent was removed by evaporation under reduced pressure. Quality control measurements using LCMS were performed on random samples of the final library. For transferring the peptides to 384 well plates, the dry peptide derivatives were dissolved in 35 µl of printing buffer and reformatted with automated liquid handling systems. Peptide microarrays were produced using a non-contact high performance microarray printer on epoxy-modified slides (PolyAn; Germany). All peptides and controls were deposited in three identical sub-arrays, enabling analysis of assay homogeneity and reliability of the results. Peptide microarrays were scanned after printing process and statistical values were generated for identification and quality control of each individual spot. Subsequently, peptide microarray surfaces were deactivated using appropriate quenching solutions, washed with water and dried using microarray centrifuges. Resulting peptide microarrays were stored at 4°C until use.
2.2. Sample selection
Thirty-six (36) serum or plasma samples were obtained from previously performed studies in the Barouch laboratory and were selected to represent a spectrum of potential preclinical and clinical uses for the microarray. Samples included (a) plasma from 5 HIV-1-infected individuals from North America with high HIV-1 viral load and 5 HIV-uninfected controls, (b) serum from 5 human subjects vaccinated with one dose of adenovirus 26 (Ad26) vaccine expressing clade A HIV-1 Env and 5 naive human controls, (c) serum from 5 rhesus monkeys vaccinated with 6 doses of clade C HIV-1 Env protein and 2 naïve monkey controls, and (d) serum from 5 guinea pigs vaccinated with 6 doses of clade C HIV-1 Env protein and 3 naïve guinea pig controls.
2.3. Microarray incubation and immunolabeling
Microarray slides were incubated with serum or plasma using the manual method, essentially as described (Masch et al., 2010). Serum or plasma was diluted 1:200 in SuperBlock T20 (TBS) Blocking Buffer (Thermo Scientific). Slides were placed in the individual chambers of a Sarstedt Quadriperm Dish and incubated in 4 mL of diluted serum/plasma for 1 hr at 30° C. Slides were then washed with 5 mL of TBS-Buffer + 0.1%Tween20 for 3 minutes on a shaker at room temperature for 5 washes. Next, slides were incubated with Alexa Fluor 647-conjugated AffiniPure Mouse Anti-Human IgG (H+L) (Jackson ImmunoResearch Laboratories) for human or monkey samples for 1 hr in the dark on a shaker at room temperature. Alexa Fluor 647-conjugated AffiniPure Goat Anti-Guinea Pig IgG (H+L) (Jackson ImmunoResearch Laboratories) was used for guinea pig samples. Slides were then washed 5 times with TBS-Buffer with 0.1%Tween20, and 5 times with deionized water. To dry, slides were placed in a 50 mL conical and spun at 1500 rpm for 5 minutes. Of note, all batches of slides were run in parallel with a control slide that is incubated with secondary antibody alone.
2.3 Microarray image analysis
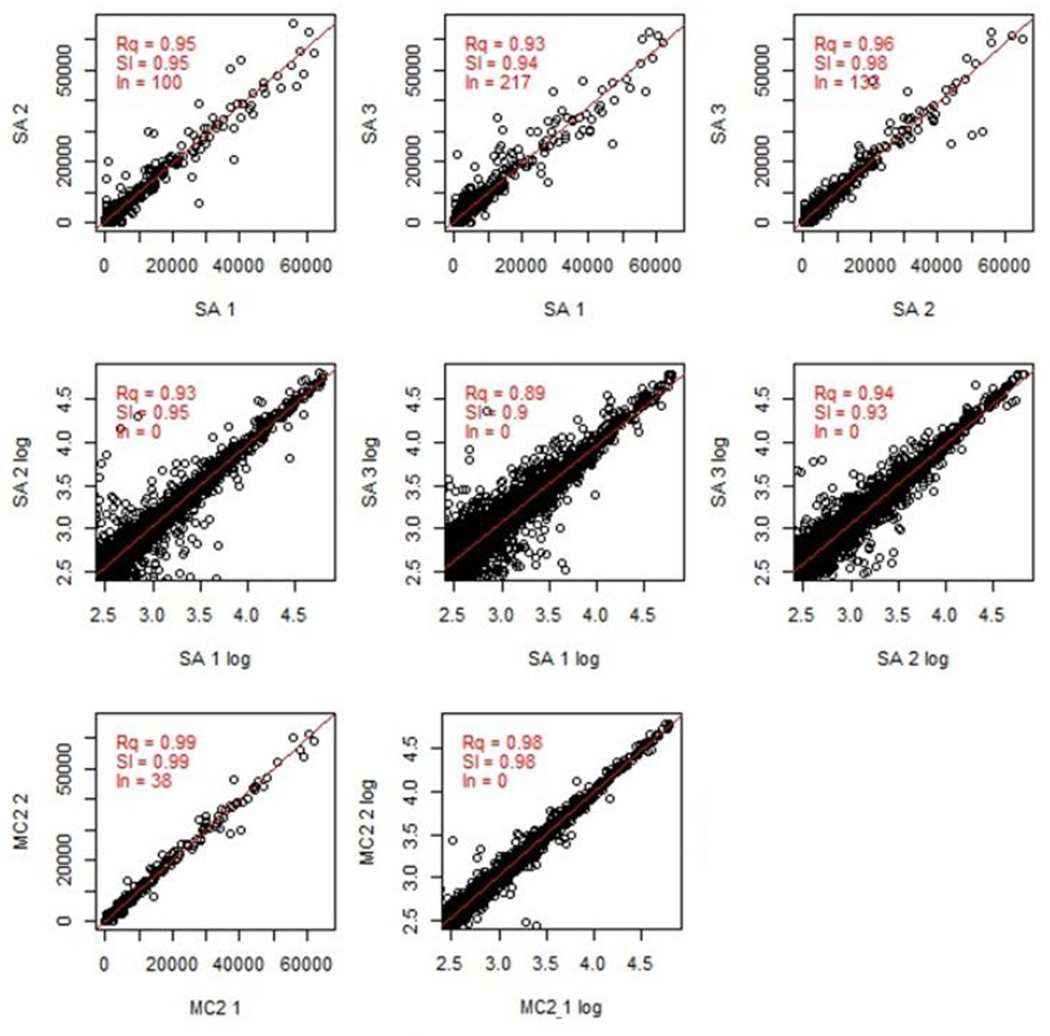
Slides were scanned with a GenePix 4300A scanner (Molecular Devices), using 635 nm and 532 nm lasers at 500 PMT and 100 Power settings. Images were saved as TIF files. The fluorescent intensity for each feature (peptide spot) was calculated using GenePix Pro 7 software and GenePix Array List (GAL) file, a text file with specific information about the location, size, and name of each feature on the slide. This analysis created a GenePix Results (GPR) file. We then calculated the mean fluorescent intensity across the triplicate sub-arrays (SAs) for each feature; if the coefficient of variation was greater than 0.5, then the mean of the two closest values was used. These calculations were performed with a custom-designed R script “MakeDat_V04” (available as Appendix 1) and R software package 2.15.2. Data was saved as a comma-delimited DAT file usable by Excel (Microsoft). MakeDat_V04 also created scatterplots of the correlation between the feature fluorescent intensities of sub-array 1 and 2, sub-array 2 and 3, and sub-array 1 and 3 as a measure of assay quality (Figure 3).
Figure 3. Quality analysis of raw microarray data.
The signal distribution of a representative microarray is displayed, as well as the correlation between slide sub-arrays. This data is following incubation with serum from an HIV-1-infected subject. SA, subarray; Rq, R squared; Sl, slope; In, intersection with the y axis; MC2, two closest values between the 3 sub-arrays; MC2 1 and MC2 2, each component of MC2.
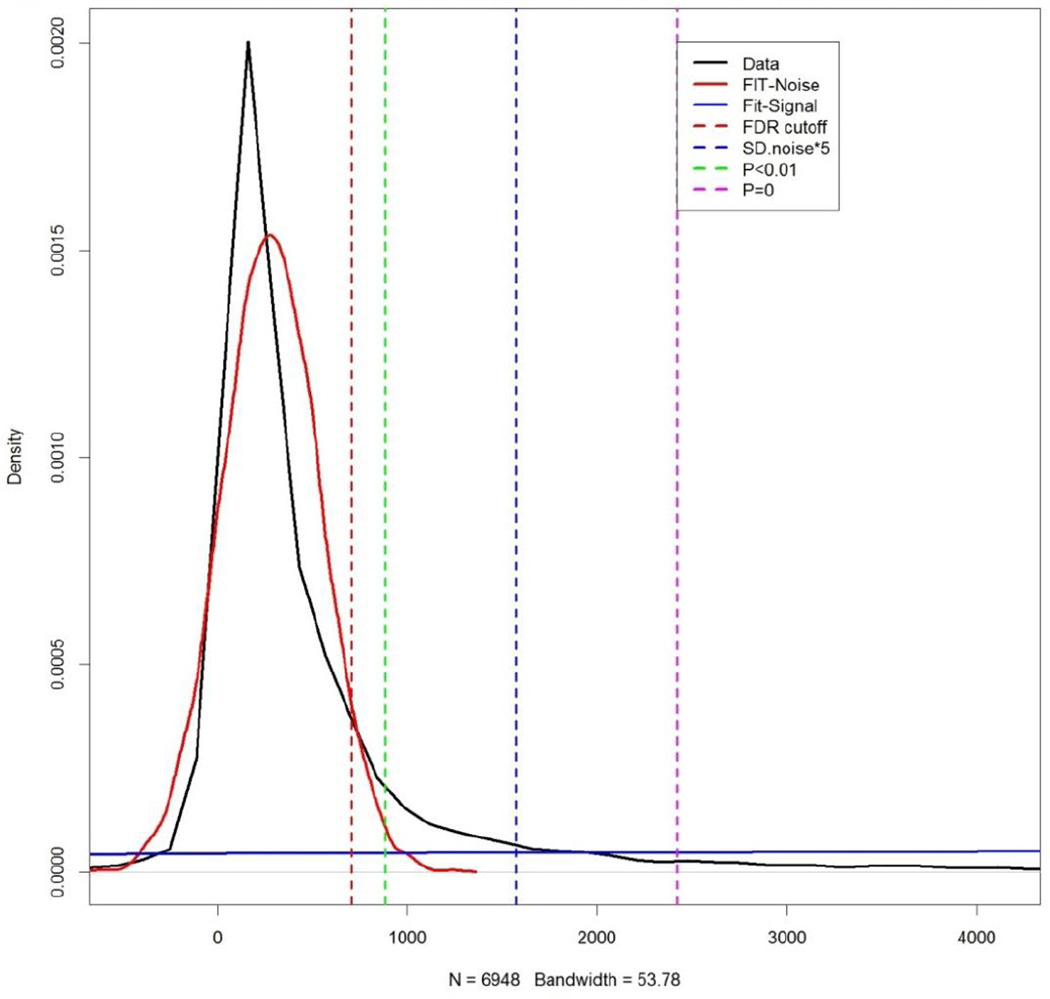
The threshold value used to define a minimum positive fluorescent intensity was calculated for each slide using the computational tool rapmad (Robust Alignment of Peptide MicroArray Data, available for free at http://tron-mainz.de/tron-facilities/computational-medicine/compmed) and the custom-designed R script “calc_THRES_KS_130711_V02” (available as Appendix 2). This analysis is based on the method as previously described (Renard et al., 2011) and distinguishes those peptide features that carry a signal from those features that only display noise. Data from each individual slide was combined with data from the control slide to create two distributions of data (noise and signal). We then calculated four potential threshold values for positivity with increasing levels of stringency: the false discovery rate cutoff (FDR cutoff), the point at which the chance that signal is noise is P<0.01, 5 standard deviations above the mean of the noise distribution (SD.noise*5), and the point at which the chance that signal is noise is very low at P<10−16.
2.4 Analysis of magnitude, breadth, and depth of antibody binding
The raw magnitude, or fluorescent intensity, of antibody binding to individual peptides (averaged over the 3 sub-arrays as described above) was sorted and categorized by (1) HIV-1 protein, (2) amino acid start position as aligned to HXB2 HIV-1 reference strain, and (3) HIV-1 clade or CRF within which the peptide sequence can be found. This sorting was performed using the custom-designed R script “Table_select_V01” (available as Appendix 3). To correct for any direct binding of the secondary antibody to linear peptides, the fluorescent intensity of antibody binding measured on the control slide was subtracted from the fluorescent intensity of antibody binding measured on the sample slide. Finally, all corrected fluorescent intensities were compared to the calculated threshold for positivity, and all values above the threshold were considered positive (with the rest of the values changed to “0” and considered negative). For these studies, we chose the threshold SD.noise*5.
To calculate the breadth of antibody binding, we evaluated the number of positive peptides for each sample and aligned the peptide sequences to eliminate overlap. If any positive peptide sequences shared 5 or more contiguous amino acids, we assumed that the peptides were recognized by the same antigen-binding site on a single antibody; these overlapping sequences were conservatively defined as a single positive “binding site.” If the first and last overlapping peptide in a string of overlapping peptides shared 4 or less amino acids, we assumed that the peptides were recognized by a minimum of two antibody sites (on either two antibodies or the same antibody). This approach to calculating antibody breadth is based on established methods to calculate T cell breadth, essentially as described in (Stephenson et al., 2012). The primary difference is that the overlapping region for T cells is usually 9 or more amino acids, reflecting the structure of CD4/CD8 T cell binding pockets. For antibodies, the antigen binding-site can range in length, and for conformational epitopes may not be contiguous. For this analysis, 5 amino acids was chosen as the minimum overlap region for antibody binding based on the fact that, apart from a handful of 3 and 4 AA sequences, 5 AA is the minimum length reported for human antibody binding to linear HIV-1 sequences as per the LANL HIV Immunology Database (Theoretical Biology and Biophysics, 2014). Conformational epitopes cannot be directly assessed with linear peptide microarray.
To calculate the depth of antibody responses, we evaluated the overlapping sequences of each binding site and determined the number of unique sequence variations of the binding site that were present. We then calculated the average number of variations/binding site for each sample. We also determined the relative frequency of clade or CRF-specific antibody responses. To do this we first defined distinct clade or CRF peptide ‘sets’ that included any peptide whose sequence had been identified in that clade or CRF (see Figure 1B). If a sequence could be found across multiple clades, it was included in multiple sets. We then calculated the percent of positive peptides within each set to provide a relative measure of clade- or CRF-specific antibody responses that could be comparable across sets of different sizes. To maximize our ability to detect differences in clade- or CRF-specific antibody responses, we restricted this analysis to the variable regions V1V2 and V3 of gp120.
3. Results and Discussion
3.1 Evaluation of coverage of global HIV-1 sequence diversity
In designing this microarray, our goal was to develop a tool to measure the diversity of HIV-1-specific antibody binding to linear HIV-1 epitopes from global sequences. To determine how well the peptide library represented global HIV-1 sequence diversity, we analyzed coverage using the program package MosaicVaccines.1.2.11 as described above. We found that the peptide library covered the majority of sequences in the Los Alamos National Database (Table 1), including gp120 (50.2 %), gp41 (65.5 %), Gag p17 (58.4 %), and Gag p24 (86.2 %). Of note, for some protein regions a small group of 15-mer peptides sufficed to span a reported antibody binding site, but because the site was of high sequence diversity with no conserved sequences, the observed coverage was low (e.g. VIF_1 with 9% coverage reported).
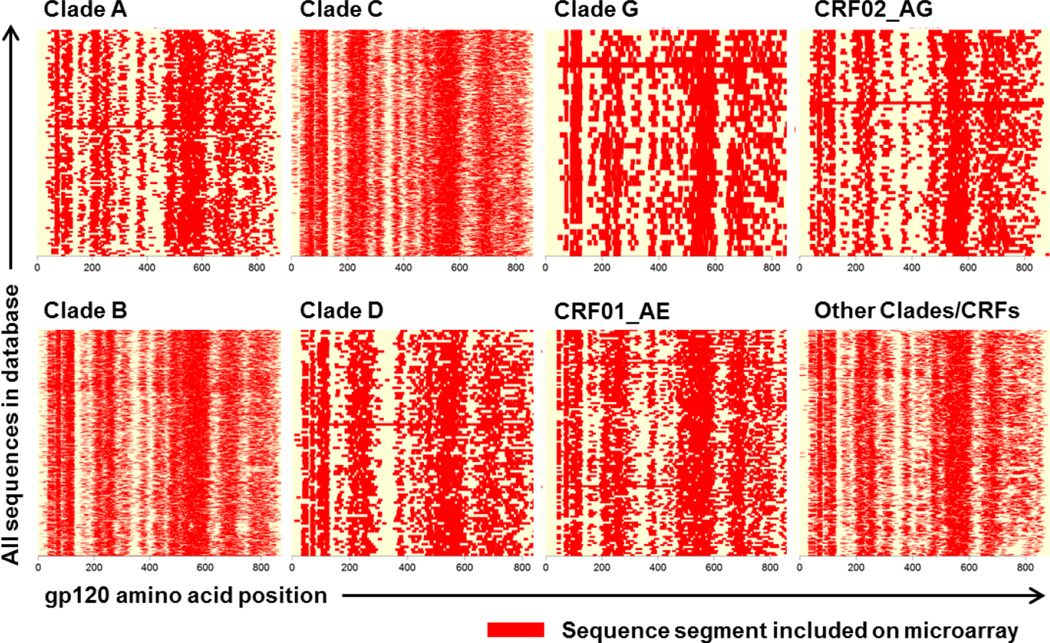
We also evaluated the coverage of gp120 sequences from clades A, B, C, D, G, CRF01_AE, CRF02_AG, and a summary population of all other clades (Figure 2). This analysis demonstrated that for each clade- or CRF-specific sequence, 50% of the sequence (on average) was covered by peptides on the microarray. As expected, in the variable regions of the HIV-1 proteome lower coverage was achieved, as for the variable loops in ENV V1/V2 (HXB2 131–196) or V4 (HXB2 385–418). However, the microarray reached a maximum of 95 peptide variants for each location within the most variable regions of HIV-1 Env, and an average of 7 peptide variants for each location on HIV-1 Env, Gag, Nef, Pol, Rev, Tat, and Vif.
Figure 2. Coverage of HIV-1 peptide library for gp120 by clade.
Top row: A, C, G, CRF02_AG; bottom row: B, D, CRF01_AE, and all other clades. The X-direction of each plot represents the sequence of gp120. Sequences segments included on the microarray are depicted in red. In the Y-direction all sequences for the respective clade from the alignment HIV1_ALL_2009_ENV_PRO.fasta are shown (total 2248). The average coverage (horizontally) for each used HIV-1 sequence is 50%. The evaluation of coverage was performed presuming the same length of all sequences for one protein or fragment within a given clade.
The diversity of linear peptides on the global HIV-1 microarray described here is in contrast to the composition of the predominant HIV-1 peptide microarray previously reported in the literature (Tomaras et al., 2011; Karasavvas et al., 2012; Gottardo et al., 2013; Imholte et al., 2013). This previous microarray includes gp160 subtype consensus sequences from six HIV-1 group M subtypes (A, B, C, D, CRF_01 and CRF_02) and a consensus group M gp160, Con-S. In contrast to the global microarray reported here, this previous microarray contains less than a quarter of the number of peptides (1,423 vs. 6,564), excludes variable sequences by design, and does not include any non-Env proteins, making it potentially less optimal for quantifying HIV-1 antibody epitope diversity.
3.2 Evaluation of microarray assay quality and determination of threshold values for positivity
Given the density of peptides on the microarray (19,692 peptides over 3 triplicate sub-arrays), we designed a program to evaluate the quality of raw microarray data following sample incubation and immunolabeling, as described above. Figure 3 demonstrates representative results of this analysis following microarray incubation with plasma from an HIV-1-infected subject. As shown in this example, the program provides a snapshot of how well the results from each sub-array correlate with each other; in this case the correlation ranged from R2=0.93 to 0.96. We also designed a program to determine a threshold value above which a signal can be considered “positive” (Renard et al., 2011). Figure 4 demonstrates representative results of this analysis when the microarray was incubated with plasma from an HIV-1-infected subject. By providing four potential threshold values with varying stringency, the program allows the user to decide whether his or her analysis will have greater sensitivity or specificity in detecting antibody binding.
Figure 4. Determination of threshold values for positivity.
The signal and noise distribution of data from a sample microarray is displayed, as well as the correlation between slide sub-arrays. Microarray data is following incubation with plasma from an HIV-1-infected subject. FDR, false discovery rate; SD, standard deviation.
3.3 Quantitating and visualizing antibody binding diversity
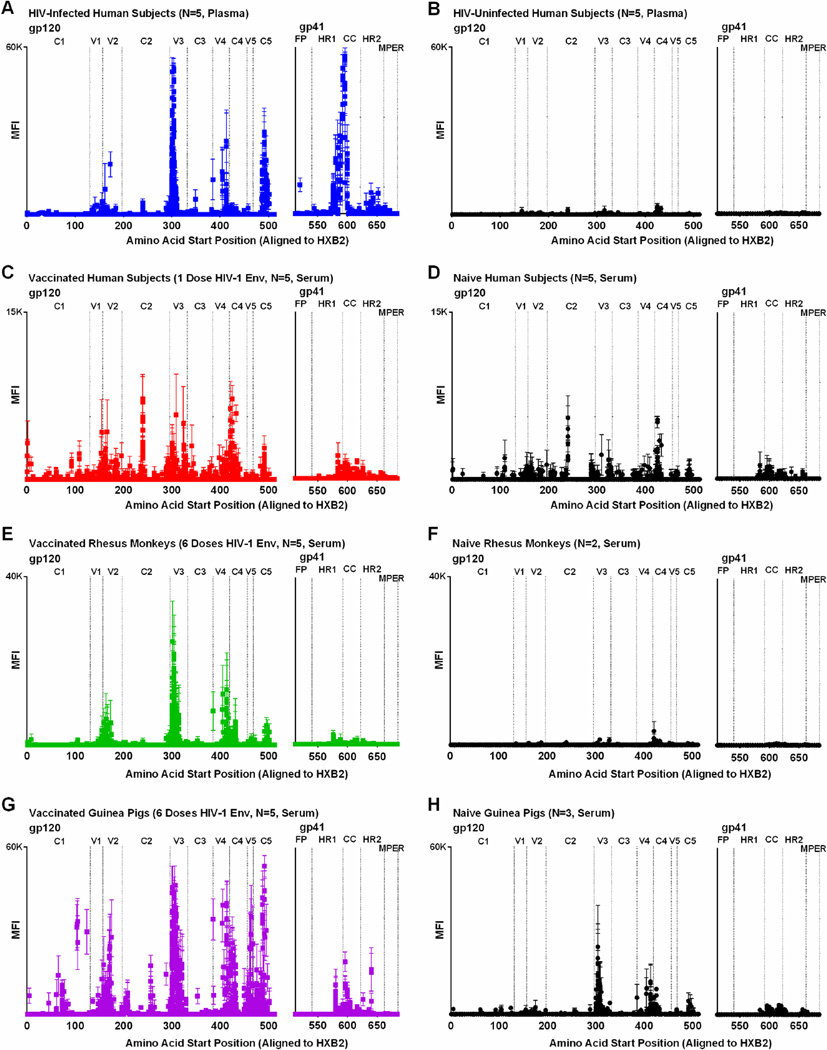
The goal of this project was to develop a method to both quantitate and visualize antibody binding patterns to diverse HIV-1 sequences for the purpose of HIV-1 vaccine and therapeutic research. To visualize binding patterns, one can plot the magnitude of peptide binding (MFI) by peptide location (starting amino acid position). For instance, Figure 5A demonstrates the gp140-specific binding pattern among HIV-1-infected subjects, where the average MFI per peptide is shown for the 5 subjects. In this example, peak MFI values were observed at the V3 region of gp120 and the CC loop region of gp41, with maximum values about 60,000 MFI, consistent with well-described immunodominant regions in HIV-1 infection (Goudsmit, 1988; Tomaras et al., 2008; Tomaras and Haynes, 2009; McMichael et al., 2010). Among HIV-uninfected controls, there were a handful of nonspecific positive peptides, but peak values did not rise above 4,500 MFI (Figure 5B). For comparison, Figure 5C shows the binding pattern among human subjects vaccinated with a single priming dose of Ad26-EnvA HIV-1 vaccine. Here peak binding values were observed to V1, V2 and V3 linear peptides, with maximum MFIs up to about 12,000. The lower MFI of vaccinees compared to HIV-1-infected subjects is expected given receipt of only one dose of vaccine without subsequent boosting, but were still above those observed in naïve controls (Figure 5D). Of note, sera from naïve controls demonstrated more nonspecific positive peptides than plasma from naïve controls.
Figure 5. IgG binding to linear HIV-1 gp140 peptides in HIV-1-infected subjects and vaccine recipients.
The mean fluorescent intensity (MFI) of positive HIV-1 gp140 peptides are plotted against the amino acid start position as aligned to HXB2 HIV-1 reference strain for (A) 5 HIV-1-infected human subjects with high viral load, (B) 5 HIV-uninfected plasma controls, (C) 5 human subjects following one dose of Ad26-EnvA HIV-1 vaccine, (D) 5 naïve human serum controls, (E) 5 rhesus monkeys following 6 doses of HIV-1 Clade C Env protein, (F) 2 naïve rhesus monkey serum controls, (G) 5 guinea pigs following 6 doses of HIV-1 Clade C Env protein, and (H) 3 naïve guinea pig serum controls. Symbols represent the average MFI from infected or vaccinated subjects (color squares) and controls (black circles); bars represent SEM.
As shown in Figure 5E–H, the peptide microarray can also be used to map antibody binding patterns in two animal models commonly used in HIV-1 vaccine research: rhesus macaques and guinea pigs (Nkolola et al., 2010; Barouch et al., 2012; Barouch et al., 2013; Nkolola et al., 2014). In both examples, animals were vaccinated with 6 serial doses of clade C HIV-1 protein and developed a similar binding pattern, with peak responses at V3. The higher MFIs among vaccinated animals compared to humans is likely due to the increased number of boosts received by the animals. Of note, naïve guinea pig samples demonstrated higher backgrounds than naïve human or monkey samples.
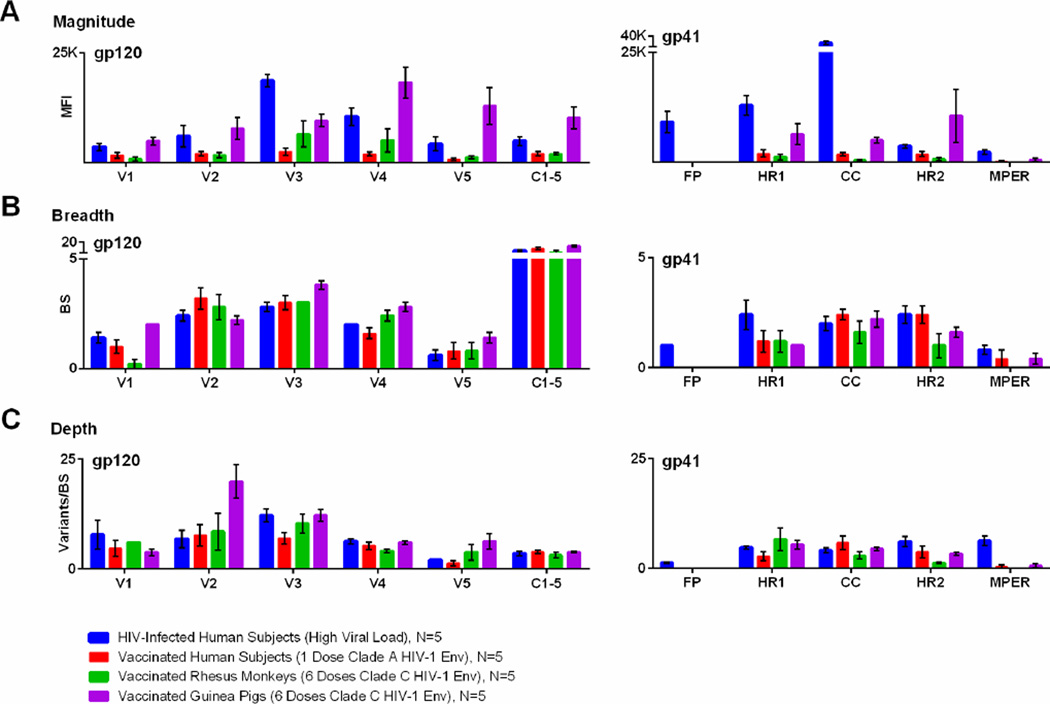
While maps of antibody binding can provide a useful tool to visualize binding patterns, they are less useful for the quantitative comparison of groups or HIV-1 regions. To provide such quantitative data, we calculated the average MFI of peptide binding sorted by region and HIV-1 protein (Figure 6A); magnitude can be compared across subjects or vaccine platforms as long as the dilution factor for the assay is kept constant, as was done in these experiments. As demonstrated in Figure 6A, the microarray can help characterize which regions of the HIV-1 envelope are preferentially targeted. For example, in HIV-1-infected subjects, V3-specific binding was significantly greater than to any other gp120 region (P<0.02 for all comparisons by t-test) and CC loop-specific binding was greater than to any other gp41 region (P<0.002 for all comparisons by t-test). In contrast, human vaccinees did not show a preference for V3 or CC loop responses, although the vaccine included these antigens.
Figure 6. Magnitude, breadth, and depth of HIV-1-specific antibody binding, as measured by global HIV-1 peptide microarray.
Antibody responses for four groups are are plotted by gp120 and gp41 region, including (A) the mean fluorescent intensity of positive peptides (magnitude), (B) the number of binding sites (breadth), and (C) the mean number of epitope variants at each binding site (depth). The four groups include HIV-1-infected subjects (blue), vaccinated human subjects (red), vaccinated rhesus monkeys (green), and vaccinated guinea pigs (purple). Bars represent SEM.
It is also useful to know whether HIV-1-specific antibodies are binding to a limited region of the HIV-1 envelope or if multiple areas are targeted. Figure 6B demonstrates the number of binding sites (“breadth”) by gp120 and gp41 region for our four groups of samples. Here, we can see that while the vaccinated human subjects had relatively low magnitude gp140 binding compared to HIV-1-infected subjects, there was no discernable difference in antibody breadth between the two groups. This ability to distinguish between magnitude and breadth is important in HIV-1 vaccine research. For example, if a particular vaccine candidate elicits low magnitude but broad antibody responses, then one might decide to change the vaccine vector or schedule to boost responses. On the other hand, if the vaccine candidate elicits high magnitude but narrow antibody responses, then one might decide to retain the same vector and schedule, but change the immunogen to broaden the specificity.
We also developed the microarray to measure the cross-clade binding of HIV-1-specific antibodies. Figure 6C demonstrates the mean number of epitope variants per binding site by gp120 and gp41 region for the four groups of samples. This analysis shows that the Ad26-EnvA vaccine elicited comparable depth of V4 binding as seen in the HIV-1-infected subjects (5 vs. 6 variants/binding site, P=NS by t-test), even though the vaccine elicited significantly lower magnitude of V4 binding (1,955 vs. 10,468 MFI, P=0.0031 by t-test). In addition, the depth of V2 binding among vaccinated guinea pigs could not be predicted by magnitude alone. For example, while HIV-1-infected humans and HIV-1-vaccinated guinea pigs had the same magnitude of V2-specific responses (5,998 vs. 7,770 MFI, P=NS by t-test), the vaccinated guinea pigs had significantly greater depth of V2-specific binding (7 vs. 20 variants/binding site, P=0.0161 by t-test). Despite substantial differences in the human and guinea pig studies, this example demonstrates how the microarray can discriminate between magnitude and depth of antibody responses. This information may be highly relevant to HIV-1 vaccine researchers who aim to design a global HIV-1 vaccine capable of blocking acquisition of diverse HIV-1 strains.
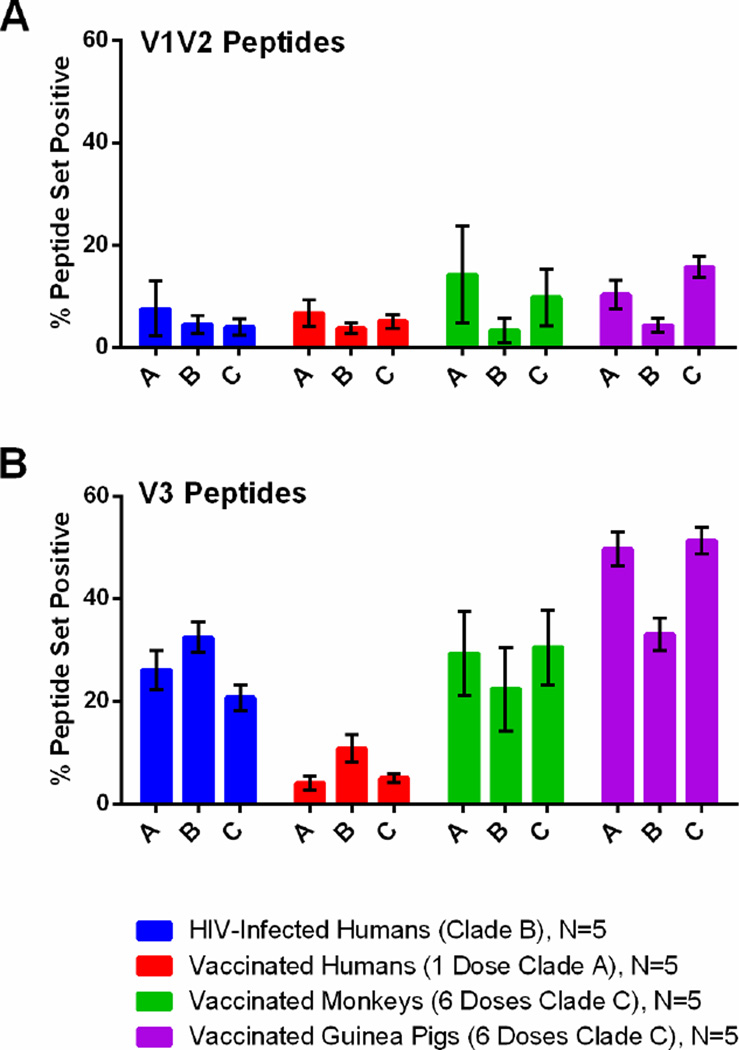
We also calculated the relative clade- or CRF-specific binding present for the three most frequent clades (A, B, and C). Figure 7 demonstrates the percent of each clade- or CRF-specific peptide set that was positive for the four groups within the variable regions V1V2 and V3. In Figure 7A, we can see that among vaccinated monkeys and guinea pigs, V1V2-specific responses were increased compared to the other cohorts, and that binding to clades A and C V1V2 peptides predominated, whereas clade B-specific binding was relatively low. This finding likely reflects the fact that both monkeys and guinea pigs received clade C Env immunogens. In contrast, in Figure 7B, we can see that among HIV-1-infected subjects, who had increased V3-specific responses, binding to clade B peptides predominated. This finding presumably reflects the fact that these subjects were from North America and were infected with clade B HIV-1. These data suggest that the microarray may not only be useful for measuring cross-clade immune responses following vaccination, but also may have an application in serotyping HIV-1-infected subjects. Further studies with larger numbers of HIV-1-infected subjects from different regions could test this hypothesis.
Figure 7. Cross-clade HIV-1 antibody binding as measured by global HIV-1 peptide microarray.
The percent of clade- or CRF-specific peptide sets that are positive for V1V2 and V3 regions of gp120. The four cohorts include HIV-1-infected subjects (blue), vaccinated human subjects (red), vaccinated rhesus monkeys (green), and vaccinated guinea pigs (purple). Bars represent SEM.
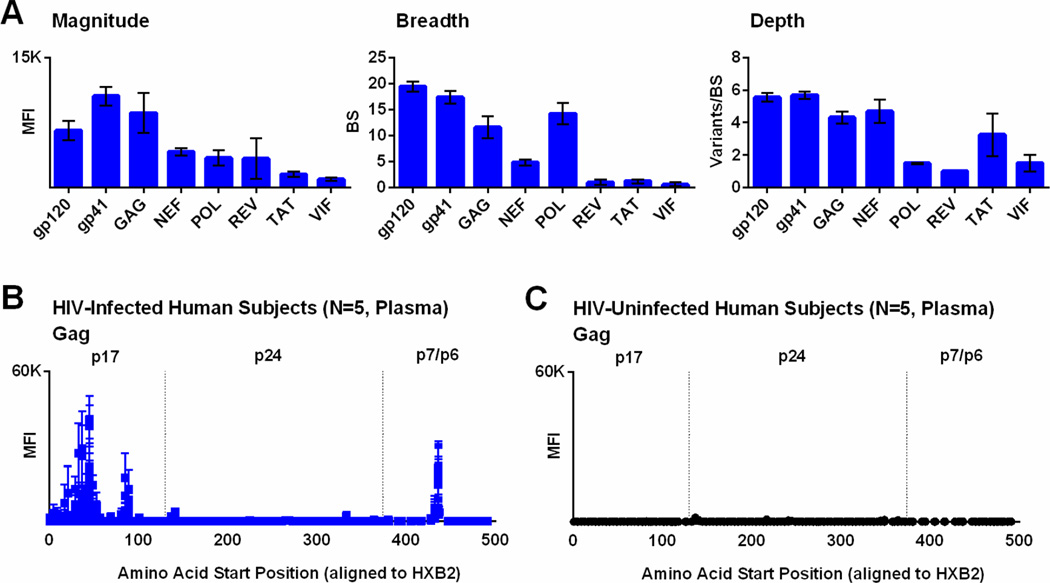
Finally, we also designed the microarray to assess HIV-1-specific binding across the HIV-1 proteome. In Figure 8A, we demonstrate the magnitude, breadth, and depth of HIV-1-specific binding to gp120, gp41, Gag, Nef, Pol, Rev, Tat, and Vif proteins among 5 HIV-1-infected human subjects. We observed that gp41 (which includes regions from the cytoplasmic tail) has the highest binding magnitude, followed by Gag. Figure 8B shows the antibody binding pattern for Gag among 5 HIV-1-infected subjects; peak values are noted within the p17 region, with very little Gag-specific binding among naïve controls (Figure 8C). Antibody binding to non-Env proteins may be relevant to evaluate vaccine potency and for certain non-neutralizing antibodies (Lewis, 2014). Some studies have also shown an association between anti-p24 responses and virologic control (Weber et al., 1987; Trkola et al., 2004). In addition, the microarray might be useful to assess vaccine-induced seroreactivity in the context of HIV-1 vaccine clinical trials.
Figure 8. IgG binding to linear HIV-1 peptides from across the HIV-1 proteome in HIV-1-infected subjects.
(A) The mean fluorescent intensity (MFI) of positive HIV-1 peptides (magnitude), number of binding sites (breadth), and mean variants per binding site (depth) are shown for multiple HIV-1 proteins for 5 HIV-1-infected human subjects. Bars represent SEM. In addition, the MFI of positive HIV-1 Gag peptides are plotted against the amino acid start position as aligned to HXB2 HIV-1 reference strain for 5 HIV-1-infected human subjects (B) and 5 HIV-uninfected controls (C). Bars represent SEM.
4. Conclusions
As more HIV-1 vaccine candidates progress into clinical trials, it is important to develop new tools to assess the epitope diversity of HIV-1-specific antibodies. Here we report the development of a global HIV-1 peptide microarray based on a library of 6,564 peptides covering the majority of sequences in the Los Alamos National Laboratory HIV-1 sequence database. This microarray provides a method to measure the magnitude, breadth, and depth of IgG binding to linear HIV-1 peptides, allowing for a more in depth analysis of antibody epitope diversity than is currently available. Such knowledge may contribute to improvements in HIV-1 vaccine design and development, or to a better understanding of immune responses to HIV-1 infection. The major limitations are that this assay does not measure conformational antibodies or antibody function. Nevertheless, when used in conjunction with other antibody assays, the microarray assays should prove useful for both preclinical and clinical HIV-1 research.
Acknowledgements
This research was supported by the National Institutes of Health (AI060354 to K.E.S.; AI078526, AI084794, AI095985, and AI096040 to D.H.B.), the Bill and Melinda Gates Foundation (OPP 1033091, OPP1040741 to D.H.B.), and the Ragon Institute of MGH, MIT, and Harvard (to K.E.S. and D.H.B.). Plasma and serum samples from human subjects were obtained from studies conducted by the AIDS Clinical Trials Group and the NIH Integrated Preclinical/Clinical AIDS Vaccine Development Program. We thank E. Rosenberg, L. Baden, M. Seaman, C. Bricault, J. Iampietro, H. Li, and Z. Kang for providing generous advice, assistance, and reagents.
Appendix 1. MakeDat_V04 R Script
Note, all provided R scripts are available under the terms of the Free Software Foundation’s GNU General Public License, as outlined at http://www.r-project.org/.
PAR_dir <- ".\\"
PAR_val <- "F635.Mean"
PAR_filter <- FALSE
PAR_flags <- c("−100") # "−50", "−75", "−100"
PAR_array <- 1 # 1 or 21
PAR_multiwell <- matrix( c(1,2,7,8,13,14,
3,4,9,10,15,16,
5,6,11,12,17,18,
19,20,25,26,31,32,
21,22,27,28,33,34,
23,24,29,30,35,36,
37,38,43,44,49,50,
39,40,45,46,51,52,
41,42,47,48,53,54,
55,56,61,62,67,68,
57,58,63,64,69,70,
59,60,65,66,71,72,
73,74,79,80,85,86,
75,76,81,82,87,88,
77,78,83,84,89,90,
91,92,97,98,103,104,
93,94,99,100,105,106,
95,96,101,102,107,108,
109,110,115,116,121,122,
111,112,117,118,123,124,
113,114,119,120,125,126) ,ncol=21)
#################################################################################################
######################
readGPR = function(name) {
headersize <- read.table(name, header=FALSE, skip=1, nrows=1)
GPR <- read.table(name, header=TRUE, skip=as.numeric(headersize[1,1])+2, sep ="\t",
strip.white=TRUE, colClasses="character")
GPR <- GPR[order(as.numeric(GPR$Column)),]
GPR <- GPR[order(as.numeric(GPR$Row)),]
GPR <- GPR[order(as.numeric(GPR$Block)),]
return(GPR)
}
getMC2 = function(val1, val2, val3) {
tmp <- cbind(rbind(abs(val1-val2),abs(val1-val3),abs(val2-
val3)),rbind(mean(c(val1,val2)), mean(c(val1,val3)),
mean(c(val2,val3))),rbind(val1,val1,val2),rbind(val2,val3,val3),rbind(1,1,2),rbind(2,3,3))
colnames(tmp) <- c("Diff", "MC2", "MC2_1", "MC2_2", "MC2_SA1", "MC2_SA2")
tmp <- tmp[order(tmp[,1]),]
return(tmp[1,])
}
getMMC2 = function(val1, val2, val3) {
val <- mean(c(val1, val2, val3))
if(sd(c(val1, val2, val3))/val > 0.5){
val <- getMC2(val1, val2, val3)["MC2"]
}
return(val)
}
filterDAT = function(dat) {
tmp <- NULL
exclude = c("AA","GGSGGGSDYKDDDDK","printingbuffer","empty")
for(i in 1:nrow(dat)) {
if( length(unlist(strsplit(dat[i,"Annotation"],"|",fixed=TRUE))) == 4 & (! (dat[i,"ID"] %in%
exclude)) ) {
tmp <- c(tmp,TRUE)
} else {
tmp <- c(tmp,FALSE)
}
}
return(dat[tmp,])
}
plotplus = function(x ,y ,name ,xl ,yl, MINVAL, MAXVAL) {
xtmp <- subset(x, !(x==-Inf|y==-Inf))
ytmp <- subset(y, !(x==-Inf|y==-Inf))
x <- xtmp
y <- ytmp
plot(x, y, main=name, xlab=xl, ylab=yl, xlim=c(MINVAL,MAXVAL), ylim=c(MINVAL,MAXVAL))
tmp1 <- lm(y~x)
abline(tmp1, col="red")
tmp2 <- summary(tmp1)
pos1 <- MAXVAL - (MAXVAL-MINVAL)/10*1
pos2 <- MAXVAL - (MAXVAL-MINVAL)/10*2
pos3 <- MAXVAL - (MAXVAL-MINVAL)/10*3
text(MINVAL,pos1,pos=4,labels=paste("Rq =",round(as.numeric(tmp2[8]),digits=2)),col="red")
text(MINVAL,pos2,pos=4,labels=paste("Sl
=",round(as.numeric(tmp1$coefficients[2]),digits=2)),col="red")
text(MINVAL,pos3,pos=4,labels=paste("In =",round(as.numeric(tmp1$coefficients[1]))),col="red")
}
#################################################################################################
######################
getDAT = function(fname,GPR,BLOCKS=NULL,INDEX=NULL){
fname <- unlist(strsplit(fname,"\\",fixed=TRUE))
fname <- fname[length(fname)]
fname <- substr(fname,1, nchar(fname)-4)
if(is.null(BLOCKS)){
ARRAY <- GPR
}else{
ARRAY <- subset(GPR, as.numeric(GPR$Block) %in% BLOCKS)
fname <- sprintf("%s_ARRAY%.2d",fname,INDEX)
}
### dat ###
SA <- NULL
SA_LINES <- nrow(ARRAY)/3
for(i in 1:3){
SA[[i]] <- ARRAY[(SA_LINES*(i-1)+1):(SA_LINES*(i)),,drop=FALSE]
}
if(FALSE %in% unique(c(SA[[1]]$Name == SA[[2]]$Name, SA[[1]]$Name == SA[[3]]$Name))){
print("ERROR")
return()
}
DAT <- cbind(SA[[1]]$ID, SA[[1]]$Name, SA[[1]]$Annotation, SA[[1]]$Flags, SA[[2]]$Flags,
SA[[3]]$Flags, SA[[1]][[PAR_val]], SA[[2]][[PAR_val]], SA[[3]][[PAR_val]])
colnames(DAT) <- c("ID", "Name", "Annotation", "SA1_Flags", "SA2_Flags", "SA3_Flags", "SA1",
"SA2", "SA3")
tmp <- apply(DAT[,c("SA1","SA2","SA3")],c(1,2),as.numeric)
DAT <- cbind(DAT, apply(tmp,1,mean), apply(tmp,1,median), apply(tmp,1,sd),
apply(tmp,1,sd)/apply(tmp,1,mean), apply(tmp,1,min), apply(tmp,1,max))
colnames(DAT)[colnames(DAT)==""] <- c("Mean", "Median", "SD", "CV", "Min", "Max")
MC2 <- NULL
MMC2 <- NULL
for(i in 1:nrow(DAT)){
MC2 <- rbind(MC2,
getMC2(as.numeric(DAT[i,"SA1"]),as.numeric(DAT[i,"SA2"]),as.numeric(DAT[i,"SA3"]))[2:4])
MMC2 <- c(MMC2,
getMMC2(as.numeric(DAT[i,"SA1"]),as.numeric(DAT[i,"SA2"]),as.numeric(DAT[i,"SA3"])))
}
DAT <- cbind(DAT, MC2, MMC2)
### corr ###
tmp[cbind(SA[[1]]$Flags %in% PAR_flags, SA[[2]]$Flags %in% PAR_flags, SA[[2]]$Flags %in%
PAR_flags)] <- NA
colnames(tmp) <- paste("CORR_",colnames(tmp),sep="")
DAT <- cbind(DAT,tmp)
DAT <- cbind(DAT, apply(tmp,1,mean,na.rm=T), apply(tmp,1,median,na.rm=T),
apply(tmp,1,sd,na.rm=T), apply(tmp,1,sd,na.rm=T)/apply(tmp,1,mean,na.rm=T),
apply(tmp,1,min,na.rm=T), apply(tmp,1,max,na.rm=T))
DAT[DAT==Inf] <- NA
DAT[DAT==-Inf] <- NA
DAT[DAT=="NaN"] <- NA
colnames(DAT)[colnames(DAT)==""] <- c("CORR_Mean", "CORR_Median", "CORR_SD", "CORR_CV",
"CORR_Min", "CORR_Max")
CORR_MC2 <- NULL
CORR_MMC2 <- NULL
for(i in 1:nrow(DAT)){
VAL <- tmp[i,!is.na(tmp[i,])]
if(length(VAL)==1){
CORR_MC2 <- rbind(CORR_MC2, c(VAL,VAL,VAL))
CORR_MMC2 <- c(CORR_MMC2, VAL)
}else if(length(VAL)==2){
CORR_MC2 <- rbind(CORR_MC2, c(mean(VAL),VAL))
CORR_MMC2 <- c(CORR_MMC2, mean(VAL))
}else if(length(VAL)==3){
CORR_MC2 <- rbind(CORR_MC2, getMC2(VAL[1],VAL[2],VAL[3])[2:4])
CORR_MMC2 <- c(CORR_MMC2, getMMC2(VAL[1],VAL[2],VAL[3]))
}else{
CORR_MC2 <- rbind(CORR_MC2, c(NA,NA,NA))
CORR_MMC2 <- c(CORR_MMC2, NA)
}
}
colnames(CORR_MC2) <- c("CORR_MC2","CORR_MC2_1","CORR_MC2_2")
DAT <- cbind(DAT, CORR_MC2, CORR_MMC2)
if(PAR_filter){
DAT <- filterDAT(DAT)
}
DAT <- DAT[order(DAT[,"Name"]),]
write.table(DAT, file=sprintf("%s.dat",fname), quote=TRUE, sep="\t", row.names=FALSE,
col.names=TRUE)
### image ###
NCOLOR <- 256
PAL <- rev(heat.colors(NCOLOR))
MINVAL <- 0
MAXVAL <- 65535
MINVAL_LOG <- 2.5
MAXVAL_LOG <- log10(MAXVAL)
PAR_valB <- "B635.Mean"
png(filename=sprintf("%s.png",fname), width=5*200, height=4*200, res=100)
layout(rbind(
c(1,3,5,6,7),
c(1,3,8,9,10),
c(2,4,11,13,15),
c(2,4,12,14,16)
))
par(mar=c(0,4,2,1))
plot(as.numeric(ARRAY$X), as.numeric(ARRAY$Y)*(−1), col=PAL[(NCOLOR-1)/(MAXVAL-
MINVAL)*(as.numeric(ARRAY[[PAR_val]])-MINVAL)+1], pch=15, cex=0.8, xlab="", ylab="", axes=FALSE,
main="Array signal", yaxs="i", xaxs="i", xlim=c(min(as.numeric(GPR$X)), max(as.numeric(GPR$X))),
ylim=c(max(as.numeric(GPR$Y))*(−1), min(as.numeric(GPR$Y))*(−1)))
tmp <- log10(as.numeric(ARRAY[[PAR_val]]))
tmp[tmp<MINVAL_LOG] <- MINVAL_LOG
plot(as.numeric(ARRAY$X), as.numeric(ARRAY$Y)*(−1), col=PAL[(NCOLOR-1)/(MAXVAL_LOG-
MINVAL_LOG)*(tmp-MINVAL_LOG)+1], pch=15, cex=0.8, xlab="", ylab="", axes=FALSE, yaxs="i",
xaxs="i", main="Array signal log", xlim=c(min(as.numeric(GPR$X)), max(as.numeric(GPR$X))),
ylim=c(max(as.numeric(GPR$Y))*(−1), min(as.numeric(GPR$Y))*(−1)))
plot(as.numeric(ARRAY$X), as.numeric(ARRAY$Y)*(−1), col=PAL[(NCOLOR-1)/(MAXVAL-
MINVAL)*(as.numeric(ARRAY[[PAR_valB]])-MINVAL)+1], pch=15, cex=0.8, xlab="", ylab="", axes=FALSE,
main="Array background", yaxs="i", xaxs="i", xlim=c(min(as.numeric(GPR$X)),
max(as.numeric(GPR$X))), ylim=c(max(as.numeric(GPR$Y))*(−1), min(as.numeric(GPR$Y))*(−1)))
tmp <- log10(as.numeric(ARRAY[[PAR_valB]]))
tmp[tmp<MINVAL_LOG] <- MINVAL_LOG
plot(as.numeric(ARRAY$X), as.numeric(ARRAY$Y)*(−1), col=PAL[(NCOLOR-1)/(MAXVAL_LOG-
MINVAL_LOG)*(tmp-MINVAL_LOG)+1], pch=15, cex=0.8, xlab="", ylab="", axes=FALSE, yaxs="i",
xaxs="i", main="Array background log", xlim=c(min(as.numeric(GPR$X)), max(as.numeric(GPR$X))),
ylim=c(max(as.numeric(GPR$Y))*(−1), min(as.numeric(GPR$Y))*(−1)))
par(mar=c(4,4,2,1))
plotplus(as.numeric(DAT[,"SA1"]),as.numeric(DAT[,"SA2"]),"",sprintf("SA %d",1),sprintf("SA
%d",2), MINVAL, MAXVAL)
plotplus(as.numeric(DAT[,"SA1"]),as.numeric(DAT[,"SA3"]),"",sprintf("SA %d",1),sprintf("SA
%d",3), MINVAL, MAXVAL)
plotplus(as.numeric(DAT[,"SA2"]),as.numeric(DAT[,"SA3"]),"",sprintf("SA %d",2),sprintf("SA
%d",3), MINVAL, MAXVAL)
plotplus(log10(as.numeric(DAT[,"SA1"])),log10(as.numeric(DAT[,"SA2"])),"",sprintf("SA %d
log",1),sprintf("SA %d log",2), MINVAL_LOG, MAXVAL_LOG)
plotplus(log10(as.numeric(DAT[,"SA1"])),log10(as.numeric(DAT[,"SA3"])),"",sprintf("SA %d
log",1),sprintf("SA %d log",3), MINVAL_LOG, MAXVAL_LOG)
plotplus(log10(as.numeric(DAT[,"SA2"])),log10(as.numeric(DAT[,"SA3"])),"",sprintf("SA %d
log",2),sprintf("SA %d log",3), MINVAL_LOG, MAXVAL_LOG)
plotplus(MC2[,"MC2_1"],MC2[,"MC2_2"],"",sprintf("MC2 %d",1),sprintf("MC2 %d",2), MINVAL,
MAXVAL)
plotplus(log10(MC2[,"MC2_1"]),log10(MC2[,"MC2_2"]),"",sprintf("MC2 %d log",1),sprintf("MC2 %d
log",2), MINVAL_LOG, MAXVAL_LOG)
plot(density(as.numeric(ARRAY[[PAR_val]])), xlim=c(MINVAL,MAXVAL), main="", xlab="Signal")
plot(density(log10(as.numeric(ARRAY[[PAR_val]]))), xlim=c(MINVAL_LOG,MAXVAL_LOG), main="",
xlab="Signal log")
tmp <- NULL
tmp[["SA1"]] <- as.numeric(DAT[,"SA1"])
tmp[["SA2"]] <- as.numeric(DAT[,"SA2"])
tmp[["SA3"]] <- as.numeric(DAT[,"SA3"])
boxplot(tmp, ylim=c(MINVAL,MAXVAL), ylab="Signal")
tmp <- NULL
tmp[["SA1"]] <- log10(as.numeric(DAT[,"SA1"]))
tmp[["SA2"]] <- log10(as.numeric(DAT[,"SA2"]))
tmp[["SA3"]] <- log10(as.numeric(DAT[,"SA3"]))
boxplot(tmp, ylim=c(MINVAL_LOG,MAXVAL_LOG), ylab="Signal log")
dev.off()
}
#################################################################################################
######################
files <- dir(path=PAR_dir, pattern="*.gpr", full.names=TRUE)
files <- files[!file.info(files)$isdir]
files <- files[substr(files,nchar(files)-3,nchar(files)) == ".gpr"]
for(i in 1:length(files)){
print(sprintf("Processing file %d out of %d",i,length(files)))
GPR <- readGPR(files[i])
if(PAR_array==1){
getDAT(files[i],GPR)
}else{
for(j in 1:PAR_array){
print(sprintf("Processing array %d out of %d",j,PAR_array))
getDAT(files[i],GPR,PAR_multiwell[,j],j)
}
}
}
Appendix 2: calc_THRES_KS_130711_V02 R Script
library(rapmad)
INDEX<-read.table("sample.xls",header=F,sep = "\t", strip.white=T,skip=1)
INDEX.dat<-INDEX[INDEX[,1]=="dat",]
if(ncol(INDEX.dat)==4){
LIST.1 <- c(as.character(INDEX.dat[,2]),as.character(INDEX.dat[,4]))
}else{
LIST.1 <- c(as.character(INDEX.dat[,2]))
}
LIST.1<-LIST.1[LIST.1!=""]
FILENAME<-"OUTFILE_THRESH.Rdat"
DAT<-NULL
for (cnt1 in 1:length(LIST.1)){
DAT.TMP<-read.table(paste(LIST.1[cnt1],sep=""),header=TRUE,sep = "\t", strip.white=T)
DAT<-cbind(DAT,DAT.TMP$MMC2)
}
#INDEX<-read.table(paste("INDEX_2070_NEW.txt",sep=""),header=TRUE,sep = "\t", strip.white=T)
CN<-substr(LIST.1,1,nchar(LIST.1)-4)
colnames(DAT)<-CN
DAT<-cbind(DAT.TMP[,c(1:3)],DAT)
save(DAT,file=FILENAME)
####Plot of Signal Distribution for Raw Data
TAB.OUT<-NULL
png(filename = paste("01_thresholds_raw_data.png",sep=""), width = 3200, height =
1600*round((length(LIST.1)+.1)/2),
units = "px", pointsize = 40, bg = "white", res = NA,
restoreConsole = TRUE)
par(mfrow=c(round((length(LIST.1)+.1)/2),2))
par(mai=c(2.5,2.5,1,.5))
for (c1 in (4:ncol(DAT))){
FIT.TMP<-signal.gmm.generic(DAT[,c1],logScale=F)
y1<-rnorm(10000,mean = FIT.TMP[[2]][1], sd = FIT.TMP[[3]][1])
y2<-rnorm(10000,mean = FIT.TMP[[2]][2], sd = FIT.TMP[[3]][2])
D<-density(DAT[,c1])
plot(density(DAT[,c1]),lwd=4,main=colnames(DAT)[c1],xlim=c(FIT.TMP[[2]][1]-
(FIT.TMP[[3]][1]*3),FIT.TMP[[2]][1]+(FIT.TMP[[3]][1]*15)))
lines(density(y1),col="red",lwd=4)
lines(density(y2),col="blue",lwd=4)
abline(v=FIT.TMP[[5]],lwd=3,col="red",lty=2)
abline(v=FIT.TMP[[2]][1]+(FIT.TMP[[3]][1]*5),lwd=3,col="blue",lty=2)
DAT.STAT<-cbind(DAT[,c1],FIT.TMP[[1]])
DAT.STAT.p0.01<-DAT.STAT[DAT.STAT[,2]<0.01,]
abline(v=min(DAT.STAT.p0.01[,1]),lwd=3,col="green",lty=2)
DAT.STAT.p0<-DAT.STAT[DAT.STAT[,2]==0,]
abline(v=min(DAT.STAT.p0[,1]),lwd=3,col="magenta",lty=2)
legend(min(DAT.STAT.p0[,1]),max(D$y),c("Data","FIT-Noise","Fit-Signal","FDR
cutoff","SD.noise*5","P<0.01","P=0"),lwd=4,
lty=c(1,1,1,2,2,2,2),
col=c("black","red","blue","red","blue","green","magenta"))
TAB.OUT<-rbind(TAB.OUT,cbind(colnames(DAT)[c1],FIT.TMP[[5]],FIT.TMP[[2]][1]+(FIT.TMP[[3]][1]*5),min(DAT.ST
AT.p0.01[,1]),min(DAT.STAT.p0[,1])))
}
dev.off()
##make DAT-CTRL
DAT.COR<-NULL
DAT.ID.KEEP<-NULL
if (ncol(INDEX.dat)==4){
INDEX.dat.corr<-INDEX.dat[INDEX.dat[,4]!="",]
for (c1 in (1:nrow(INDEX.dat.corr))){
DAT.ID.TMP<-
substr(as.character(INDEX.dat.corr[c1,2]),1,nchar(as.character(INDEX.dat.corr[c1,2]))-4)
DAT.ID.KEEP<-c(DAT.ID.KEEP,DAT.ID.TMP)
CTR.ID.TMP<-
substr(as.character(INDEX.dat.corr[c1,4]),1,nchar(as.character(INDEX.dat.corr[c1,4]))-4)
DAT.COR<-cbind(DAT.COR,DAT[[DAT.ID.TMP]]-DAT[[CTR.ID.TMP]])
}
colnames(DAT.COR)<-paste(DAT.ID.KEEP,"COR",sep="_")
DAT.COR<-cbind(DAT[,1:3],DAT.COR)
####Plot of Signal Distribution for Corrected Data
png(filename = paste("02_thresholds_corrected_data.png",sep=""), width = 3200, height =
1600*round((length(DAT.ID.KEEP)+.1)/2),
units = "px", pointsize = 40, bg = "white", res = NA,
restoreConsole = TRUE)
par(mfrow=c(round((length(DAT.ID.KEEP)+.1)/2),2))
par(mai=c(2.5,2.5,1,.5))
for (c1 in (4:ncol(DAT.COR))){
FIT.TMP<-signal.gmm.generic(DAT.COR[,c1],logScale=F)
y1<-rnorm(10000,mean = FIT.TMP[[2]][1], sd = FIT.TMP[[3]][1])
y2<-rnorm(10000,mean = FIT.TMP[[2]][2], sd = FIT.TMP[[3]][2])
D<-density(DAT.COR[,c1])
plot(density(DAT.COR[,c1]),lwd=4,main=colnames(DAT.COR)[c1],xlim=c(FIT.TMP[[2]][1]-
(FIT.TMP[[3]][1]*3),FIT.TMP[[2]][1]+(FIT.TMP[[3]][1]*15)))
lines(density(y1),col="red",lwd=4)
lines(density(y2),col="blue",lwd=4)
abline(v=FIT.TMP[[5]],lwd=3,col="red",lty=2)
abline(v=FIT.TMP[[2]][1]+(FIT.TMP[[3]][1]*5),lwd=3,col="blue",lty=2)
DAT.STAT<-cbind(DAT.COR[,c1],FIT.TMP[[1]])
DAT.STAT.p0.01<-DAT.STAT[DAT.STAT[,2]<0.01,]
abline(v=min(DAT.STAT.p0.01[,1]),lwd=3,col="green",lty=2)
DAT.STAT.p0<-DAT.STAT[DAT.STAT[,2]==0,]
abline(v=min(DAT.STAT.p0[,1]),lwd=3,col="magenta",lty=2)
legend(min(DAT.STAT.p0[,1]),max(D$y),c("Data","FIT-Noise","Fit-Signal","FDR
cutoff","SD.noise*5","P<0.01","P=0"),lwd=4,
lty=c(1,1,1,2,2,2,2),
col=c("black","red","blue","red","blue","green","magenta"))
TAB.OUT<-
rbind(TAB.OUT,cbind(colnames(DAT.COR)[c1],FIT.TMP[[5]],FIT.TMP[[2]][1]+(FIT.TMP[[3]][1]*5),min(DA T.STAT.p0.01[,1]),min(DAT.STAT.p0[,1])))
}
dev.off()
}
colnames(TAB.OUT)<-c("Experiment","FDR-cutoff","SD.noise*5","P<0.01","P=0")
write.table(TAB.OUT,file="thresholds.xls",quote=F,row.names=F,col.names=T,sep="\t")
Appendix 3: Table_select_V01 R script
readDAT = function(name) {
return(read.table(name, header=TRUE, sep ="\t", strip.white=TRUE, colClasses="character"))
}
filterDAT = function(dat) {
tmp <- NULL
exclude =
c("AA","empty","GGSGGGSDYKDDDDK","GGSDYKDDDDK","DYKDDDDK","printingbuffer","mouse_IgG","human_IgG
","IgG-mouse","IgG-human")
for(i in 1:nrow(dat)) {
if( length(unlist(strsplit(dat$Annotation[i],"|",fixed=TRUE))) == 4 & (!(dat$ID[i] %in%
exclude)) ) {
tmp <- c(tmp,TRUE)
} else {
tmp <- c(tmp,FALSE)
}
}
return(dat[tmp,])
}
#################################################################################################
######################
VAL <- "MMC2"
files <- read.table("samples.xls", header=FALSE, sep ="\t", strip.white=TRUE,
colClasses="character")
TAB <- NULL
for(i in 1:nrow(files)) {
print(i)
print(files[[1]][i])
DAT <- filterDAT(readDAT(files[[1]][i]))
TAB <- cbind(TAB, as.numeric(DAT[[VAL]]))
}
colnames(TAB) <- files[[2]]
rownames(TAB) <- sprintf("%s - %s", DAT$ID, DAT$Name)
TAB <- TAB[order(as.numeric(substr(rownames(TAB),19,22))),]
TAB <- apply(TAB,c(1,2),round)
OUT <- rownames(TAB)
OUT <- strsplit(OUT, " - ", fixed=T)
OUT <- do.call("rbind",OUT)
OUT <- cbind(OUT, do.call("rbind",strsplit(substr(OUT[,2],6,14),"",fixed=T))) # Protein
OUT <- cbind(OUT, as.numeric(substr(OUT[,2],16,19))+1 ) # Position+1
OUT <- cbind(OUT, do.call("rbind",strsplit(substr(OUT[,2],21,182),"",fixed=T))) # Clade
PROT <- toupper(c("env", "gag", "nef", "pol", "rev", "tat", "vif", "vpr", "vpu"))
CLADE <- c("A","A1","A2","B","C","D","F1","F2","G","H","J","K","01_AE","02_AG","03_AB","04_cpx","05_DF","0
6_cpx","07_BC","08_BC","09_cpx","10_CD","11_cpx","12_BF","13_cpx","14_BG","15_01B","16_A2D","17_B
F","18_cpx","19_cpx","20_BG","21_A2D","22_01A1","23_BG","24_BG","25_cpx","26_AU","27_cpx","28_BF"
,"29_BF","31_BC","32_06A1","33_01B","34_01B","35_AD","36_cpx","37_cpx","38_BF1","39_BF","40_BF","
42_BF","43_02G","44_BF","45_cpx","46_BF","47_BF","49_cpx","0102A","01A1","01ADF2","01AF2U","01B",
"01BC","01C","01DU","0206","0209","0213","0225","02A","02A1","02A1U","02AG","02B","02C","02D","02
GK","02U","06A1","0708","09A","13U","1819","26C","26CU","A1A2CD","A1A2D","A1C","A1CD","A1CDGKU","
A1CG","A1D","A1DHK","A1F2","A1G","A1GHU","A1GJ","A1GU","A1H","A1U","A2C","A2CD","A2D","AC","ACD",
"AD","ADGU","ADU","AF2","AF2G","AGKU","AGU","AHJU","AKU","AU","BC","BCF1","BCU","BF","BF1","BG","
CD","CF1","CF1U","CU","DF","DF1G","DG","DO","DU","F2KU","GKU","JKU","P","A3","0107","01GHJKU","11
A1","23A1","A1B","A1DK","A2G","ACFG","AG","AGH","BFG","DF1","GH","F","01F2","01U","02H","07B","11
C","CGU","x","01A1G","02F2","KU","A1A2","GJ")
colnames(OUT) <- c("Sequence","Name",PROT,"Position+1",CLADE)
OUT <- cbind(OUT, TAB)
write.table(OUT, file="table_select.xls", quote=TRUE, sep="\t", row.names=F, col.names=TRUE)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
K.E.S., G.H.N., B.T.K., and D.H.B. have no competing interests. U.R., N.P., T.K., and J.Z. are employees of JPT Peptide Technologies. N.P. is currently an employee of Bayer HealthCare Pharmaceuticals. JPT has developed, manufactures, and sells peptide microarrays similar to those used in this study. K.E.S., G.H.N., B.T.K., and D.H.B. did not receive any compensation from JPT and do not benefit in any way from the sale of JPT products including peptide microarrays.
References
- Barouch DH, Korber B. HIV-1 vaccine development after Step. Annual Reviews in Medicine. 2009;61:2.1–2.15. doi: 10.1146/annurev.med.042508.093728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, Iampietro MJ, SanMiguel A, Seaman MS, Ferrari G, Forthal DN, Ourmanov I, Hirsch VM, Carville A, Mansfield KG, Stablein D, Pau MG, Schuitemaker H, Sadoff JC, Billings EA, Rao M, Robb ML, Kim JH, Marovich MA, Goudsmit J, Michael NL. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, O’Brien KL, Simmons NL, King SL, Abbink P, Maxfield LF, Sun YH, La Porte A, Riggs AM, Lynch DM, Clark SL, Backus K, Perry JR, Seaman MS, al e. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nature Medicine. 2010;16:319–323. doi: 10.1038/nm.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, Liu J, Abbink P, Maxfield LF, Seaman MS, Dugast AS, Alter G, Ferguson M, Li W, Earl PL, Moss B, Giorgi EE, Szinger JJ, Eller LA, Billings EA, Rao M, Tovanabutra S, Sanders-Buell E, Weijtens M, Pau MG, Schuitemaker H, Robb ML, Kim JH, Korber BT, Michael NL. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. 2013;155:531–539. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerecedo I, Zamora J, Shreffler WG, Lin J, Bardina L, Dieguez MC, Wang J, Muriel A, de la Hoz B, Sampson HA. Mapping of the IgE and IgG4 sequential epitopes of milk allergens with a peptide microarray-based immunoassay. The Journal of allergy and clinical immunology. 2008;122:589–594. doi: 10.1016/j.jaci.2008.06.040. [DOI] [PubMed] [Google Scholar]
- Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, al e. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nature Medicine. 2007a;13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, Kuiken C, Haynes B, Letvin NL, Walker BD, Hahn BH, Korber BT. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007b;13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- Gaseitsiwe S, Valentini D, Mahdavifar S, Magalhaes I, Hoft DF, Zerweck J, Schutkowski M, Andersson J, Reilly M, Maeurer MJ. Pattern recognition in pulmonary tuberculosis defined by high content peptide microarray chip analysis representing 61 proteins from M. tuberculosis. PLoS One. 2008;3:e3840. doi: 10.1371/journal.pone.0003840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, Tomaras GD, Turk E, Imholte G, Eckler L, Wenschuh H, Zerweck J, Greene K, Gao H, Berman PW, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Robb ML, Michael NL, Kim JH, Zolla-Pazner S, Haynes BF, Mascola JR, Self S, Gilbert P, Montefiori DC. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS One. 2013;8:e75665. doi: 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudsmit J. Immunodominant B-cell epitopes of the HIV-1 envelope recognized by infected and immunized hosts. AIDS. 1988;2(Suppl 1):S41–S45. doi: 10.1097/00002030-198800001-00006. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans Montefiori DT, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinte RA, Fong Y, Janes H, DeCamp A, Huang Y, Rao M. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. New England Journal of Medicine. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distributions of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- Imholte GC, Sauteraud R, Korber B, Bailer RT, Turk ET, Shen X, Tomaras GD, Mascola JR, Koup RA, Montefiori DC, Gottardo R. A computational framework for the analysis of peptide microarray antibody binding data with application to HIV vaccine profiling. J Immunol Methods. 2013;395:1–13. doi: 10.1016/j.jim.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasavvas N, Billings E, Rao M, Williams C, Zolla-Pazner S, Bailer RT, Koup RA, Madnote S, Arworn D, Shen X, Tomaras GD, Currier JR, Jiang M, Magaret C, Andrews C, Gottardo R, Gilbert P, Cardozo TJ, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Paris R, Greene K, Gao H, Gurunathan S, Tartaglia J, Sinangil F, Korber BT, Montefiori DC, Mascola JR, Robb ML, Haynes BF, Ngauy V, Michael NL, Kim JH, de Souza MS, Collaboration MT. The Thai Phase III HIV Type 1 Vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res Hum Retroviruses. 2012;28:1444–1457. doi: 10.1089/aid.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber BT, Foley B, Gaschen B, Kuiken C. Retroviral Immune Response and Restoration. Totowa, New Jersey: Humana Press; 2001. Epidemiological and immunological implications of the global variability of HIV-1; pp. 1–32. [Google Scholar]
- Korber BT, Letvin NL, Haynes BF. T-cell vaccine strategies for human immunodeficiency virus, the virus with a thousand faces. Journal of virology. 2009;83:8300–8314. doi: 10.1128/JVI.00114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GK. Role of Fc-mediated antibody function in protective immunity against HIV-1. Immunology. 2014;142:46–57. doi: 10.1111/imm.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz P, Kreutzer M, Zerweck J, Schutkowski M, Thiesen HJ. Probing the epitope signatures of IgG antibodies in human serum from patients with autoimmune disease. Methods in molecular biology. 2009;524:247–258. doi: 10.1007/978-1-59745-450-6_18. [DOI] [PubMed] [Google Scholar]
- Masch A, Zerweck J, Reimer U, Wenschuh H, Schutkowski M. Antibody signatures defined by high-content peptide microarray analysis. Methods Mol Biol. 2010;669:161–172. doi: 10.1007/978-1-60761-845-4_13. [DOI] [PubMed] [Google Scholar]
- McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol. 2010;10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahtman T, Jernberg A, Mahdavifar S, Zerweck J, Schutkowski M, Maeurer M, Reilly M. Validation of peptide epitope microarray experiments and extraction of quality data. Journal of immunological methods. 2007;328:1–13. doi: 10.1016/j.jim.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Ndung’u T, Weiss RA. On HIV diversity. AIDS. 2012;26:1255–1260. doi: 10.1097/QAD.0b013e32835461b5. [DOI] [PubMed] [Google Scholar]
- Nkolola JP, Bricault CA, Cheung A, Shields J, Perry J, Kovacs JM, Giorgi E, van Winsen M, Apetri A, Brinkman-van der Linden EC, Chen B, Korber B, Seaman MS, Barouch DH. Characterization and immunogenicity of a novel mosaic M HIV-1 gp140 trimer. J Virol. 2014;88:9538–9552. doi: 10.1128/JVI.01739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkolola JP, Peng H, Settembre EC, Freeman M, Grandpre LE, Devoy C, Lynch DM, La Porte A, Simmons NL, Bradley R, Montefiori DC, Seaman MS, Chen B, Barouch DH. Breadth of neutralizing antibodies elicited by stable, homogeneous clade A and clade C HIV-1 gp140 envelope trimers in guinea pigs. J Virol. 2010;84:3270–3279. doi: 10.1128/JVI.02252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picker LJ, Hansen SG, Lifson JD. New paradigms for HIV/AIDS vaccine development. Annual Review of Medicine. 2012;63:95–111. doi: 10.1146/annurev-med-042010-085643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard BY, Löwer M, Kühne Y, Reimer U, Rothermel A, Türeci O, Castle JC, Sahin U. rapmad: Robust analysis of peptide microarray data. BMC Bioinformatics. 2011;12:324. doi: 10.1186/1471-2105-12-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. HIV-1 nomenclature proposal. Science. 2000;288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- Stephenson KE, Barouch DH. A global approach to HIV-1 vaccine development. Immunol Rev. 2013;254:295–304. doi: 10.1111/imr.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson KE, SanMiguel A, Simmons NL, Smith K, Lewis MG, Szinger JJ, Korber B, Barouch DH. Full-length HIV-1 immunogens induce greater magnitude and comparable breadth of T lymphocyte responses to conserved HIV-1 regions compared with conserved-region-only HIV-1 immunogens in rhesus monkeys. J Virol. 2012;86:11434–11440. doi: 10.1128/JVI.01779-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Sobieszczyk ME, E MF, Hammer SM. The challenge of HIV-1 subtype diversity. New England Journal of Medicine. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theoretical Biology and Biophysics, L.A.N.L. HIV Sequence Compendium. 2009. [Google Scholar]
- Theoretical Biology and Biophysics, L.A.N.L. HIV Molecular Immunology: Maps of Ab Epitope Locations Plotted by Protein. 2010. [Google Scholar]
- Theoretical Biology and Biophysics, L.A.N.L. HIV Molecular Immunology: Antibody Epitope Summary. 2014. [Google Scholar]
- Thurmond J, Yoon H, Juiken C, Yusim K, Perkins S, Theiler J, al e. Web-based design and evaluation of T-cell vaccine candidates. Bioinformatics. 2008a;24:1639–1640. doi: 10.1093/bioinformatics/btn251. [DOI] [PubMed] [Google Scholar]
- Thurmond J, Yoon H, Kuiken C, Yusim K, Perkins S, Theiler J, Bhattacharya T, Korber B, Fischer W. Web-based design and evaluation of T-cell vaccine candidates. Bioinformatics. 2008b;24:1639–1640. doi: 10.1093/bioinformatics/btn251. [DOI] [PubMed] [Google Scholar]
- Tomaras GD, Binley JM, Gray ES, Crooks ET, Osawa K, Moore PL, Tumba N, Tong T, Shen X, Yates NL, Decker J, Wibmer CK, Gao F, Alam SM, Easterbrook P, Abdool Karim S, Kamanga G, Crump JA, Cohen M, Shaw GM, Mascola JR, Haynes BF, Montefiori DC, Morris L. Polyclonal B cell responses to conserved neutralization epitopes in a subset of HIV-1-infected individuals. J Virol. 2011;85:11502–11519. doi: 10.1128/JVI.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, Haynes BF. HIV-1-specific antibody responses during acute and chronic HIV-1 infection. Curr Opin HIV AIDS. 2009;4:373–379. doi: 10.1097/COH.0b013e32832f00c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, Cohen M, Eron J, Hicks CB, Liao HX, Self SG, Landucci G, Forthal DN, Weinhold KJ, Keele BF, Hahn BH, Greenberg ML, Morris L, Karim SS, Blattner WA, Montefiori DC, Shaw GM, Perelson AS, Haynes BF. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Kuster H, Leemann C, Oxenius A, Fagard C, Furrer H, Battegay M, Vernazza P, Bernasconi E, Weber R, Hirschel B, Bonhoeffer S, Gunthard HF, Swiss HIVCS. Humoral immunity to HIV-1: kinetics of antibody responses in chronic infection reflects capacity of immune system to improve viral set point. Blood. 2004;104:1784–1792. doi: 10.1182/blood-2004-01-0251. [DOI] [PubMed] [Google Scholar]
- Walker BD, Ahmed R, Plotkin S. Use both arms to beat HIV. Nature Medicine. 2011;17:1194–1195. doi: 10.1038/nm.2529. [DOI] [PubMed] [Google Scholar]
- Weber JN, Clapham PR, Weiss RA, Parker D, Roberts C, Duncan J, Weller I, Carne C, Tedder RS, Pinching AJ. Human immunodeficiency virus infection in two cohorts of homosexual men: neutralising sera and association of anti-gag antibody with prognosis. Lancet. 1987;1:119–122. doi: 10.1016/s0140-6736(87)91964-7. [DOI] [PubMed] [Google Scholar]
- Zhang M, Foley B, Schultz AK, Macke JP, Bulla I, Stanke M, Morgenstern B, Korber B, Leitner T. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology. 2010;7:25. doi: 10.1186/1742-4690-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]