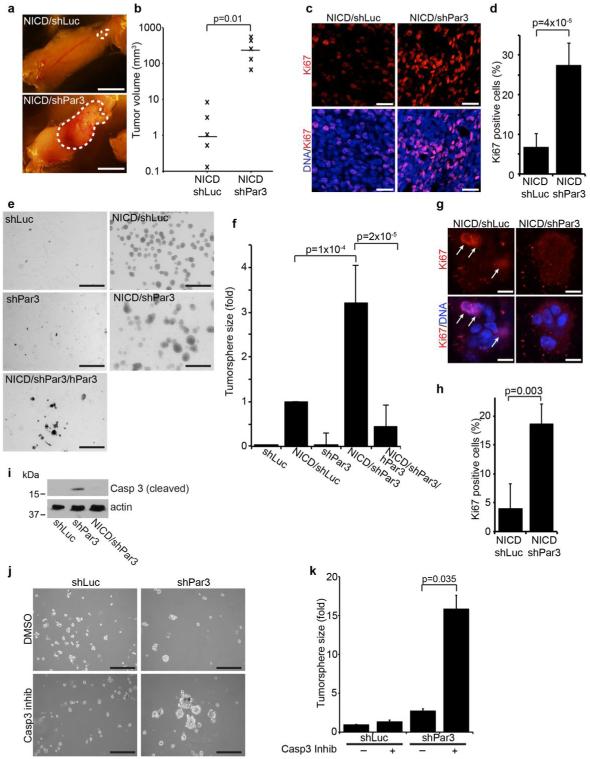
Figure 1. Loss of Par3 cooperates with NICD to promote mammary tumor growth.
(a) Mammary tumors from mammary epithelial cells expressing NICD/shLuc (control) or NICD/shPar3 (shRNA sequence: shPar3 5’-acaagcgtggcatgatcca). Primary mammary epithelial cells (MECs) were isolated from wild-type mice, infected with lentivirus to express NICD and shRNA, then transplanted into the cleared mammary fat pad. (b) Volumes of NICD/shLuc and NICD/shPar3 tumors were calculated using the formula V=□/6×(length×width2). (c) Tumor sections were immunostained for Ki67 (1:100, Abcam). (d) Quantification of cells with nuclear Ki67 from (c). Two regions from each of 3 tumors were analyzed for each. (e) MECs were infected with lentivirus to express shLuc, NICD/shLuc, shPar3, NICD/shPar3, or NICD/shPar3/hPar3 and were grown in suspension cultures for 9 days to form tumorspheres. hPar3 is resistant to the mouse-specific Par3 shRNA. (f) Quantification of tumorsphere sizes from (d). The area of each tumorsphere was measured using a custom ImageJ macro. Each independent experiment contained at least 30 tumorspheres. (g) Tumorspheres were grown for 4 days, then immunostained for Ki67. Arrows show Ki67 positive cells. (h) Quantification of cells with nuclear Ki67 from (g); n= 4 independent experiments. (i) Cell lysates were prepared from tumorspheres and immunoblotted for cleaved Caspase-3 (1:1000, Cell Signaling Technology) or actin (1:1000, Sigma) as a loading control. (j) MECs expressing shLuc or shPar3 were treated with DMSO (control) or 10μM Caspase-3 inhibitor (Ac-DEVD-CHO, BD Biosciences), and were grown for 9 days in suspension culture. (k) Quantification of tumorspheres in (j). n=3 independent samples, unless otherwise noted. Error bars=sd. p-values were calculated using the student’s t-test (b, d, h) or the ANOVA with Tukey HSD post-hoc test (f, k) using SPSS software. Scale bars=1 cm (a); 20μm (c); 1 mm (e); 200μm (j).