Figure 5.
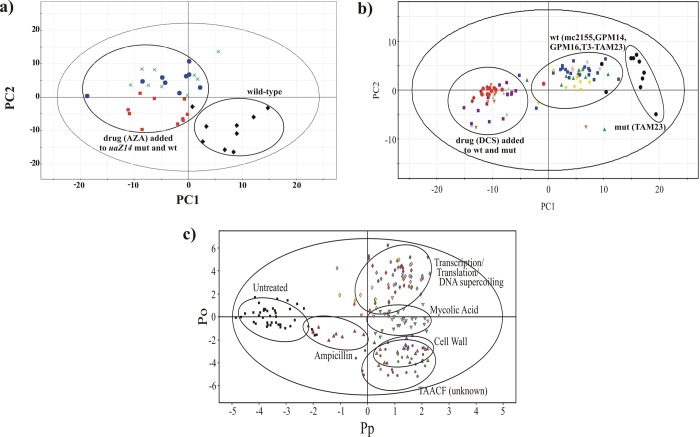
(a) The PCA scores plot comparing A. nidulansuaZ14 mutant (green X’s), wild-type with AZA (solid red squares), uaZ14 mutant with AZA (solid blue circles), and wild-type cells (solid black diamonds). The clustering pattern is comparable to the hypothetical PCA scores plot depicted in Figure 4B, which is consistent with an active and selective inhibitor. Abbreviations correspond to mutant cells (mut), wild-type cells (wt), and 8-azaxanthine (AZA). Reprinted with permission from ref (95). Copyright 2006 American Chemical Society. (b) PCA scores plot comparing Mycobacterium smegmatis mc2155 (solid blue squares), TAM23 (solid black circles), GPM14 (solid red diamonds), GPM16 (solid green upright triangles), TAM23 pTAMU3 (solid gold inverted triangles), mc2155 with DCS (solid purple squares), and TAM23 with DCS (solid red circles), GPM14 with DCS (solid purple diamonds), GPM16 with DCS (solid cyan upright triangles), and TAM23 pTAMU3 with DCS (solid orange inverted triangles). The clustering pattern is comparable to the hypothetical PCA scores plot depicted in Figure 4C, which is consistent with an active and nonselective inhibitor. Abbreviations correspond to mutant cells (mut), wild-type cells (wt) and d-cycloserine (DCS). Reprinted with permission from ref (96). Copyright 2007 American Chemical Society. (c) 2D OPLS-DA scores plot demonstrating the clustering pattern for 12 antibiotics with known biological targets and three compounds of unknown in vivo activity: untreated M. smegmatis cells (solid black squares), chloramphenicol (solid cyan diamonds), ciprofloxacin (solid gold diamonds), gentamicin (solid pink diamonds), kanamycin (solid purple diamonds), rifampicin (solid red diamonds), streptomycin (solid yellow diamonds), ethambutol (solid light-green inverted triangles), ethionamide (solid cyan inverted triangles), isoniazid (solid pink inverted triangles), ampicillin (solid red upright triangles), d-cycloserine (solid upright purple triangles), vancomycin (solid upright orange triangles), amiodorone (solid purple circles), chlorprothixene (solid light-green circles), and clofazimine (solid red circles) treated M. smegmatis cells. The ellipses correspond to the 95% confidence limits from a normal distribution for each cluster. The untreated M. smegmatis cells (solid black squares) was designated the control class, and the remainder of the cells were designated as treated. The OPLS-DA used one predictive component and six orthogonal components to yield a R2X of 0.715, R2Y of 0.803, Q2 of 0.671, and a CV-ANOVA p-value of 1.54 × 10–34. Reprinted with permission from ref (97). Copyright 2012 American Chemical Society.