Figure 9.
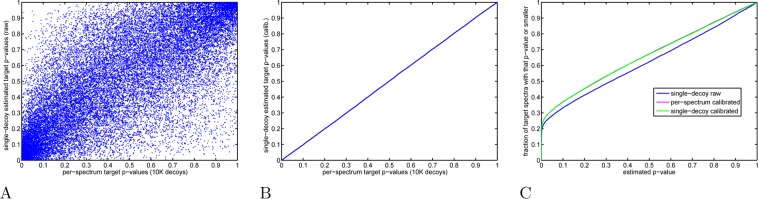
Decoy-estimated p-values: calibrated versus raw scores. The target PSM p-values were estimated from XCorr scores in three different ways: (i) p-values were estimated using the same 10K decoys per spectrum that we use to define our calibrated scores or (ii, iii) a single-decoy set was arbitrarily chosen and used to estimate the target PSM p-value according to Käll’s procedure, but once using the raw scores for both decoy and target PSMs and another time using the calibrated scores for both. (A) Scatter plot of raw score p-values (yeast, charge 2 set) computed using Käll’s procedure (a single-decoy set, y axis) and using spectrum-specific distributions generated from 10K decoys (x axis). (B) Similar to panel A, but computed using calibrated rather than raw scores. (C) The figure plots, as a function of p-value threshold, the fraction of target PSMs with p-values less than or equal to the threshold. For any nominal p-value t ≤ 0.95, we find fewer target PSMs whose raw score estimated p-value is ≤ t (blue) than when the same p-value is estimated using the calibrated scores with either the single-decoy or per-spectrum method (magenta and green, essentially in perfect agreement).
