Fig. 3. Sample purification of the E. coli RNAP.
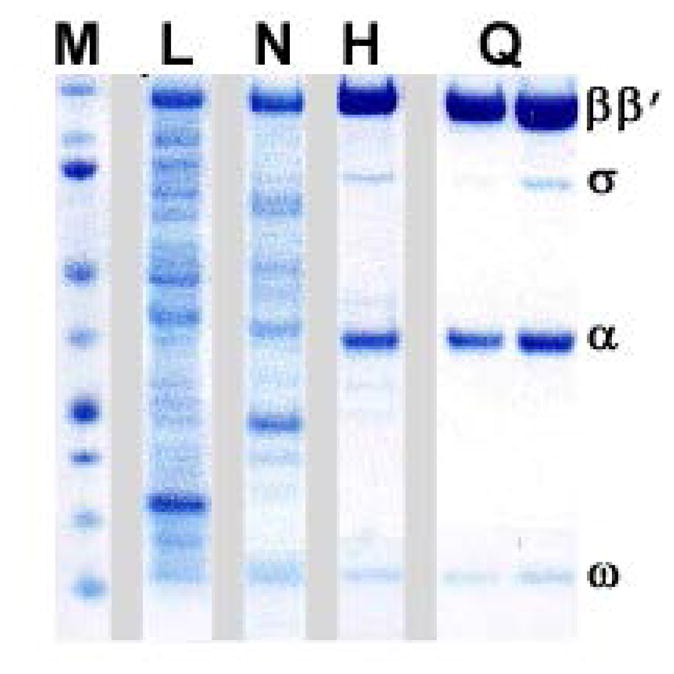
A variant with a deletion in the β′ jaw (β′ residues 1149–1190) was used. Samples of the cell lysate (L), Ni-NTA eluate (N; see section 4.3), heparin peak fractions (H; see section 4.4), and MonoQ peak fractions (H; see section 4.5) were analyzed by SDS-PAGE using a 4–12% NuPAGE® Bis-Tris pre-cast polyacrylamide gel (Life Technologies). Chromatography through the MonoQ column separates the core enzyme (left) from the holoenzyme (right). The lane marked M was loaded with the molecular weight markers; the positions of the RNAP subunits are indicated on the right.
