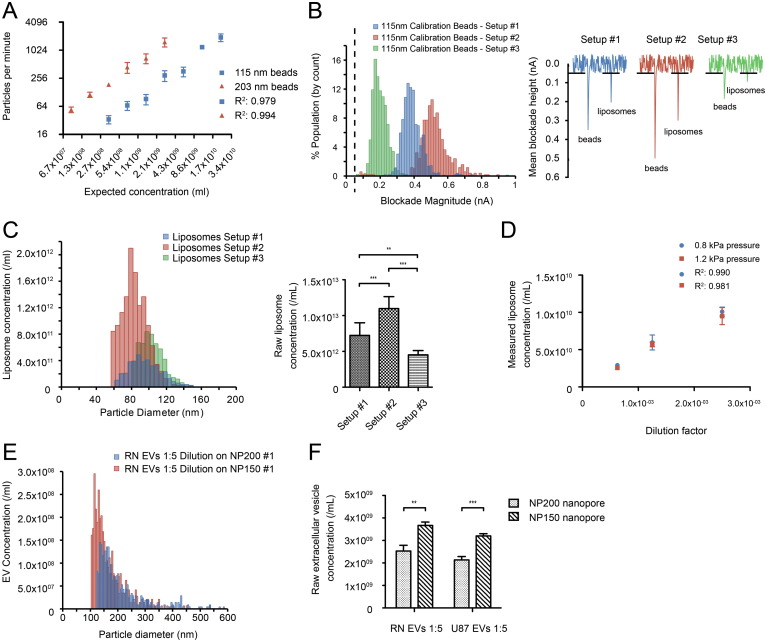
Fig. 2.
tRPS-based particle quantification. (A) Quantification of 115 and 203 nm polystyrene calibration beads. As tRPS quantification is based on the conversion of observed particle per minute counts to that of polystyrene calibration beads of known concentration, the read-out is displayed as “particles per minute”. (B) Three tRPS setups displaying the observed blockade heights for the same 115 nm calibration beads (left-panel). The dashed line illustrates the detection threshold (both panels). Reconstruction of the recorded data for beads and liposomes at the three different setups (right-panel), illustrating that the lower detection limit is the highest for setup #3, followed by setups #1 and #2. Bin size 15 pA. (C) Representative liposome size-distributions obtained at the three different tRPS setups (left-panel). For each of the three setups the measured concentration was corrected for the dilution factor to obtain raw concentration estimations (n = 6) (right-panel). Bin size 5 nm. (D) Quantification of serially diluted liposomes at two different pressure levels (n = 3). (E) Representative size-distribution obtained for RN-derived EVs on an NP200 nanopore setup and an N150A nanopore. Bin size 5 nm. (F) Raw particle concentrations were determined for RN and U87-MG derived EVs at both the NP200 and NP150 nanopore setups (n = 3). Data are mean ± s.d.