Figure 1. Structures of the caged Ang-II analogues and competitive displacement of [125I]Ang-II by caged Ang-II analogues.
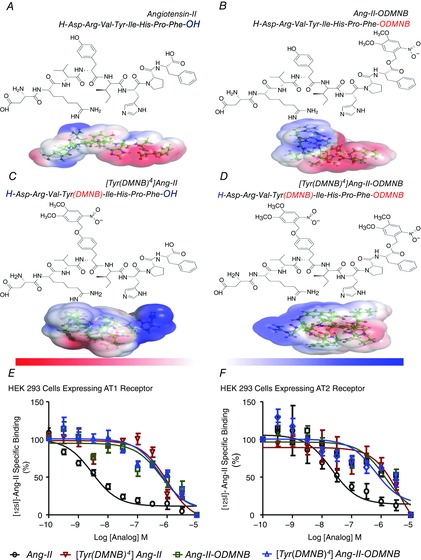
A, Ang-II. B, Ang-II-ODMNB. C, [Tyr(DMNB)4]Ang-II. D, [Tyr(DMNB)4]Ang-II-ODMNB. The position of the photosensitive DMNB moiety is indicated in brackets and was added either on the side chain of the tyrosine at position 4, the C-terminal carboxylic function of phenylalanine-8, or at both sites. Red and blue surfaces describe negative and positive electrostatic potentials (−3.5 kBT, +3.5 kBT), respectively. Three-dimensional structures were generated using PyMol visualization software. The electrostatic potentials were calculated using the Adaptive Poisson-Boltzmann Solver with the PyMol tool. (E, F, competitive displacement of [125I]Ang-II by Ang-II or caged Ang-II analogues (n = 6/condition) in HEK 293 cells transfected with AT1R or AT2R. Data are percentage of specific radioligand binding in absence of competitors. Non-specific binding was determined in the presence of 1 μm Ang-II.
