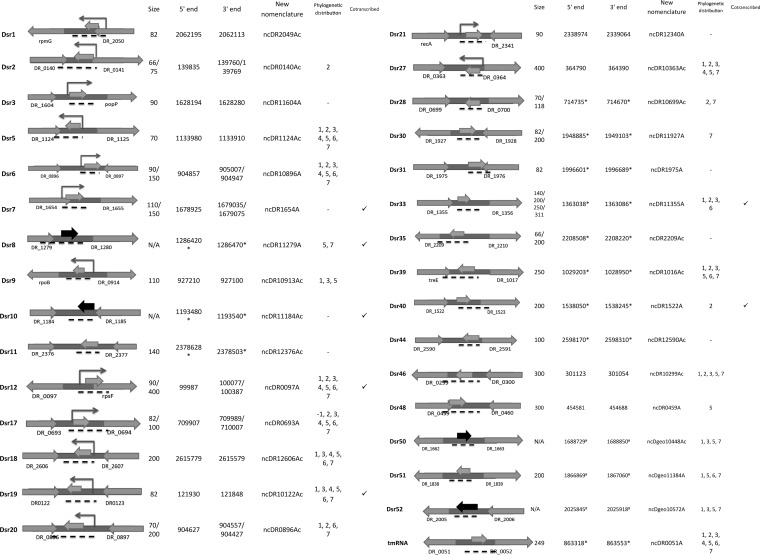
FIG 1.
Novel small RNA candidates in D. radiodurans confirmed by Northern blotting and/or RT-PCR deep sequencing. The angled arrows show the transcription start site confirmed by deep 5′ RACE. The middle arrows show where the Northern probes bind or where the PCR primers amplified. The gray middle arrows represent sRNAs that were identified by Northern blotting, and the black middle arrows represent sRNAs that identified with RT-PCR but not Northern blotting. The arrows on the sides are the annotated flanking protein-encoding regions. The black dashed lines indicate the estimated sRNA coding loci. The size of the RNAs was estimated by comparison to a phiX174 ladder. The 5′ ends were either identified by 5′ RACE or estimated by using deep sequencing data (indicated by asterisks). The 3′ ends are estimated with 5′-end coordinates and RNA size determined by Northern analysis or deep sequencing. The phylogenetic distribution shows the species for which homologous small RNAs were found: 1, Deinococcus gobiensis; 2, Deinococcus proteolyticus; 3, Deinococcus deserti; 4, Deinococcus peraridilitoris; 5, Deinococcus geothermalis; 6, Deinococcus maricopensis; 7, Deinococcus swuensis. The cotranscribed sRNAs that were identified by RT-PCR are marked in the far-right column.