FIG 1.
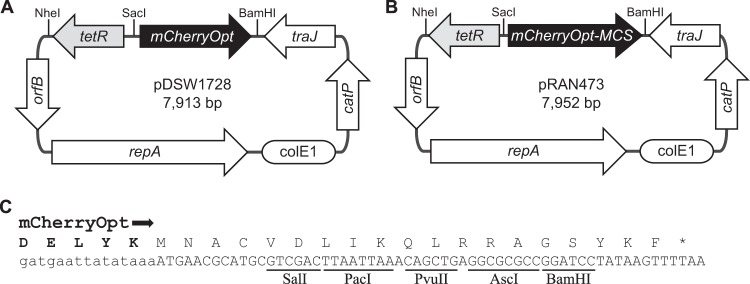
Genetic maps of C. difficile mCherryOpt plasmids. (A) pDSW1728, for studies of gene expression. Expression of mCherryOpt is under Ptet control, but the promoter-regulatory region can be replaced using the NheI and SacI restriction sites. (B) pRAN473, for studies of protein localization. (C) Multicloning site (MCS) in pRAN473. The last five amino acids derived from mCherryOpt are shown in boldface above the DNA sequence. Both plasmids are derivatives of pRPF185. Features depicted: mCherryOpt or mCherryOpt-MCS encodes mCherryOpt protein codon optimized for expression in low-GC bacteria; tetR encodes the Tet repressor from Tn10; orfB and repA, the replication region; ColE1 ori, replication region of the E. coli plasmid pBR322 modified for higher copy number; catP, the chloramphenicol acetyltransferase gene from Clostridium perfringens, conferring resistance to thiamphenicol in C. difficile or chloramphenicol in E. coli; traJ, encodes a conjugation transfer protein from plasmid RP4.
