Abstract
Reverse transcription (RT)-PCR-based virus detection from water samples is occasionally hampered by organic substances that are coconcentrated during virus concentration procedures. To characterize these organic substances, samples containing commercially available humic acid, which is known to inhibit RT-PCR, and river water samples were subjected to adsorption-elution-based virus concentration using an electronegative membrane. In this study, the samples before, during, and after the concentration were analyzed in terms of organic properties and virus detection efficiencies. Two out of the three humic acid solutions resulted in RT-quantitative PCR (qPCR) inhibition that caused >3-log10-unit underestimation of spiked poliovirus. Over 60% of the organics contained in the two solutions were recovered in the concentrate, while over 60% of the organics in the uninhibited solution were lost during the concentration process. River water concentrates also caused inhibition of RT-qPCR. Organic concentrations in the river water samples increased by 2.3 to 3.9 times after the virus concentration procedure. The inhibitory samples contained organic fractions in the 10- to 100-kDa size range, which are suspected to be RT-PCR inhibitors. According to excitation-emission matrices, humic acid-like and protein-like fractions were also recovered from river water concentrates, but these fractions did not seem to affect virus detection. Our findings reveal that detailed organic analyses are effective in characterizing inhibitory substances.
INTRODUCTION
Human enteric viruses are etiological agents that can cause clinical symptoms, such as diarrhea and vomiting. Feces and vomit from infected individuals contain a substantial amount of viruses that contaminate the water environment. Consumption of improperly treated drinking water or contaminated environmental water during recreational activity leads to waterborne infections (1, 2).
Quantitative detection of viruses present in water has been carried out worldwide in studies on the fate of viruses in the environment and potential viral infection risk (3–5). The detection of viruses in a water sample is commonly carried out using a PCR assay following virus concentration and nucleic acid extraction steps due to its superior rapidity, specificity, and sensitivity. However, the reliability of the PCR-based assay occasionally has come into question, as substances present in original samples or additives used during sample processing interfere with the nucleic acid extraction and reverse transcription (RT)-PCR (6–8). Even though several virus concentration methods have been developed, none of them can exclude the inhibitory substances completely (9).
Polyvalent cations and some organic substances, such as beef extract constituents and humic acids, are known to inhibit RT-PCR (8, 10). Often, the virus detection efficiency is determined by spiking a known amount of viruses or nucleic acids in a sample and recovering them (6, 7, 11). In the worst case, the concentrations of viruses were underestimated by 3 to 4 log10 units due to the interference (3, 7). To overcome the RT-PCR inhibition problems, a virus concentration method based on adsorption-elution with an electronegative membrane has been developed and used widely (7, 11–14). This method can exclude polyvalent cations and does not require beef extract. However, recent studies have shown that the method also results in low virus detection efficiencies, especially when a large volume of sample is processed (7, 11, 13). Our previous study found that a solution of a commercially available humic acid causes (RT) PCR inhibition after the virus concentration and nucleic acid extraction steps (7). This result suggests that certain organics in the original water samples, which are concentrated along with viruses, inhibit (RT) PCR.
Several studies have tried to keep RT-PCR efficiency high even in the presence of inhibitory substances. Additives, such as bovine serum albumin (BSA) and T4 gene 32 protein, have been shown to be effective in reducing RT-PCR inhibition (15, 16). Sample purification techniques, such as gel chromatography, cation-exchange resins, and DAX-8 resins, have been reported to be effective (17, 18). Further, sample dilution technique has been used to improve detection efficiency (3, 6). However, these methods have some limitations and are not always sufficiently effective (8, 15). Substances that hamper virus detection using RT-PCR vary depending on the quality of water samples and the virus concentration methods used. Therefore, it is necessary to characterize inhibitory substances by investigating water samples with different water qualities and virus concentrates to find an effective way to improve the virus detection efficiency. Because organic substances are the most likely causes of RT-PCR inhibition, the properties of the organic substances recovered in virus concentrates must be studied.
Humic acid is a dissolved organic fraction that is ubiquitous in natural water and is a known inhibitor of RT-PCR (8, 10, 16). Humic acid is defined as a fraction that adsorbs to an appropriate hydrophobic resin under a pH of 2, is eluted by solutions with strongly alkaline pH, and is precipitated by lowering the pH to 1 (19). The International Humic Substances Society (IHSS) uses XAD-8, a hydrophobic resin which has properties similar to DAX-8 in terms of being an adsorbent, to isolate humic acid. Even though the structure of humic acid is variable, when it is isolated by the IHSS method, its molecular size is around 1.0 kDa, and it generates a peak at a fixed location on excitation-emission matrix (EEM) spectra (20–22). Because humic substances, including humic acid, absorb UV light at 254 nm or 260 nm, the UV absorbance and the ratio between the UV absorbance and the dissolved organic carbon concentration are used as indicators of humic substances present in the natural water environment (23).
This study aimed to determine what kinds of organics are concentrated during a virus concentration step and consequently interfere with virus detection by RT-PCR. Water samples containing commercially available humic acids and river water samples were concentrated by the adsorption-elution method using an electronegative membrane, their organic contents were analyzed thoroughly, and virus detection efficiencies were determined.
MATERIALS AND METHODS
Viruses.
Poliovirus type 1 (strain LSc 2ab Sabin) (PV) was propagated in buffalo green monkey (BGM) kidney cells grown in Eagle's minimum essential medium (Nussui Seiyaku, Tokyo, Japan) containing 1% fetal bovine serum (Nichirei Corporation, Tokyo, Japan), 2.0 mM l-glutamine (Life Technologies, Tokyo, Japan), 1% antibiotic-antimycotic (Life Technologies), and 0.15% sodium bicarbonate (Life Technologies). PV was inoculated into the BGM cells and incubated for 3 days at 37°C with 5% CO2. Then, the PV was separated from the cells by freeze-thawing three times. The separated PV was purified by membrane filtration with a PTFE membrane filter (0.20-μm pore size; 25-mm diameter; Advantec, Tokyo, Japan) and gel filtration with an Illustra Microspin S-300 HR Column (GE Healthcare, Tokyo, Japan). The concentration of the purified PV (9.0 × 106 genomic copies [GC]/μl) was determined by RNA extraction followed by RT-quantitative PCR (qPCR).
Raw-sample preparation and collection.
To prepare the humic acid solutions, three commercially available humic acid powders were used: Ald-HA (Sigma-Aldrich Tokyo, Japan), Wa-HA (Wako Chemicals, Tokyo, Japan), and IH-HA (IHSS, St. Paul, MN, USA). Among the humic acid powders, IH-HA was isolated from the Suwannee River according to the IHSS method using XAD-8 resin. First, 100 mg of each powder was dissolved in 1 M NaOH solution and neutralized by HCl. Then, the solutions were diluted with Milli-Q water to appropriate concentrations (27.7 to 39.6 mg of carbon [mgC]/liter) to obtain “raw-HA samples.” Apart from the samples prepared with commercially available humic acids, three river water samples (R1 to R3) were collected from the Kyukitakami River in Miyagi Prefecture, Japan, from February to March 2012. The river water samples were kept cool until use. Here, these original river water samples are denoted “raw-river-water samples.”
Virus concentration procedures.
The raw-HA samples and the raw-river-water samples were concentrated using the adsorption-elution method with an electronegative membrane (14, 24) (Fig. 1). Two types of electronegative membranes were used; a flat-type membrane was used for a small volume (10 ml) of the raw-HA samples, and a cartridge-type membrane was used for a large volume (30 liters) of the raw-river-water samples. Because both membranes are made from the same material (i.e., mixed cellulose with ∼0.5-μm pores), the adsorption of virus to the membrane is expected to take place in the same manner. The cartridge-type membrane possesses a larger filtration area (0.1 m2) than the flat type (0.007 m2), and the cartridge type can concentrate a larger volume of samples.
FIG 1.
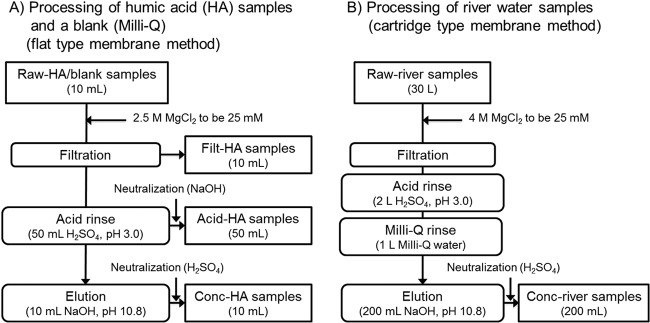
Schematic view of the sample concentration methods. Humic acid samples (Wa-HA, Ald-HA, and IH-HA) (10 ml) and a blank (Milli-Q) were concentrated using a flat-type electronegative membrane to obtain 10 ml of concentrates (conc-HA samples). During the process, filt-HA samples and acid-HA samples were collected. Thirty liters of river water samples (R1 to R3) were concentrated using a cartridge-type electronegative membrane to obtain 200 ml of concentrates (conc-river-water samples).
For raw-HA samples, 100 ml of each sample was passed through a flat-type electronegative membrane (45-mm diameter; 0.45-μm pore size; type HA; Millipore, Tokyo, Japan) after adding 2.5 M MgCl2 to a final concentration of 25 mM (filtration step) (Fig. 1A). The recovered filtrate is referred to here as the “filt-HA sample.” Then, 50 ml of 0.5 mM H2SO4 (pH 3.0) was passed through the membrane, and this filtrate was recovered in a tube containing 1 ml of 1.0 mM NaOH (pH 10.8). The recovered filtrate from this step (acid rinse step) is referred to as the “acid-HA sample,” Subsequently, 10 ml of 1.0 mM NaOH (pH 10.8) was passed through the membrane (elution step). The eluate was recovered in a tube containing 50 μl of 0.5 mM H2SO4 (pH 3.0). The recovered eluate from this step is referred to as the “conc-HA sample.” Milli-Q water was also subjected to the same adsorption-elution method as a blank test. The virus concentration process was conducted three times each for all raw-HA samples and a blank.
The raw-river-water samples were concentrated by a method using a cartridge-type membrane, which is a modification of the method using a flat-type membrane (Fig. 1B). Approximately 30 liters of the raw-river-water sample was suction filtered through a cartridge-type electronegative membrane (0.1-m2 area; 0.5-μm pore size; Opticap XL 2; Millipore, Tokyo, Japan) directly from the river using a sterilized hose and an aspirator (AS-01; As One, Osaka, Japan) (filtration step). During the filtration process, 4 M MgCl2 was sequentially dripped into the hose 1 m upstream of the filter. Then, the membrane cartridges were kept cool and transported to the laboratory. Subsequently, 2 liters of 0.5 mM H2SO4 (pH 3.0) and 1 liter of Milli-Q water were sequentially passed through the membrane (acid rinse and Milli-Q rinse steps, respectively). Finally, 200 ml of 1.0 mM NaOH (pH 10.8) was passed through the membrane. The eluate was recovered in a tube containing 1 ml of 0.5 mM H2SO4 (pH 3.0) and labeled the “conc-river sample” (elution step). The conc-river sample was further concentrated using an ultrafiltration device (Centricon Plus-70; Millipore) according to the manufacturer's protocol to obtain a final volume of 0.8 ml. The obtained samples, namely, raw-HA/raw-river, filt-HA, acid-HA, and conc-HA/conc-river samples, were spiked with the purified PV (9.0 × 106 GC/μl) to final concentrations of 9.0 × 104 GC/μl and were subjected to RT-qPCR after RNA extraction. The raw-IH-HA sample and the filt-IH-HA sample were also spiked with extracted PV RNA (9.0 × 106 GC/μl) to final concentrations of 4.5 × 104 GC/μl and were directly subjected to RT-qPCR.
Detection of PV by RT-qPCR.
The virus detection efficiency of each sample was evaluated by spiking and recovering PV by RT-qPCR using a primer pair and a TaqMan probe that are broadly reactive to enteroviruses (EVs), including PV (14). Briefly, 140 μl of the sample was spiked with 1.4 μl of PV (9.0 × 106 GC/μl; a final concentration of 9.0 × 104 GC/μl) and subjected to RNA extraction using a QiaAmp Viral RNA minikit (Qiagen) according to the manufacturer's protocol to obtain a 60-μl RNA extract. The RNA extract was subjected to the RT reaction using a High Capacity cDNA reverse transcription kit (Applied Biosystems, Tokyo, Japan). A 10-μl reaction mixture was prepared by mixing 5 μl of the RNA extract with 1 μl of 10× reverse transcription buffer (Applied Biosystems), 25 units of MultiScribe Reverse Transcriptase (Applied Biosystems), 0.4 μl of 25× deoxynucleoside triphosphates (dNTPs) (Applied Biosystems), 10 units of RNase inhibitor (Applied Biosystems), and 0.25 μl of 10 μM antisense primer (5′-ACC GGA TGG CCA ATC CAA-3′). The RT reaction was conducted with a GeneAmp PCR System 9600 (Applied Biosystems) by incubating the reaction mixture for 10 min at 25°C, 120 min at 37°C, and 5 min at 85°C. TaqMan-based qPCR was performed using a 25-μl reaction mixture that contained 5 μl of the cDNA, 12.5 μl of TaqMan Gene Expression master mix (Applied Biosystems), 1 μl each of 10 μM sense and antisense primers (5′-CCT CCG GCC CCT GAA TG-3′ and 5′-ACC GGA TGG CCA ATC CAA-3′, respectively), and 0.5 μl of 5 μM TaqMan probe (6-carboxyfluorescein [FAM]-CCG ACT ACT TTG GGT GTC CGT GTT TC- 6-carboxytetramethylrhodamine [TAMRA]). The ABI Sequence Detection System 7500 (Applied Biosystems) was used for PCR amplification with cycling conditions as follows: 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. Amplification data were collected and analyzed using Sequence Detector software version 1.3 (Applied Biosystems). To obtain a calibration curve, 10-fold serial dilutions (concentrations ranged from 1.0 × 101 to 1.0 × 106 copies per qPCR mixture) of a plasmid DNA containing the target sequence were amplified.
Determination of virus detection efficiency.
Virus detection efficiencies were calculated as the ratio of the observed spiked PV concentration in the blank (Milli-Q) and that in each virus-spiked sample. If the virus detection efficiency was lower than 10%, the efficiency was determined to be substantially low. In cases where there was substantially low virus detection efficiency, the RNA extract was diluted 100-fold with nuclease-free water to relieve RT-qPCR inhibition and subjected to RT-qPCR again.
Analysis of DOC and 254-nm UV absorbance.
Dissolved organic carbon (DOC) was measured by TOC-V (Shimadzu, Kyoto, Japan). To obtain a calibration curve, C8H4K2O4 solutions at 20, 10, 5.0, 2.0, 1.0, and 0.5 mgC/liter were used. The 254-nm-wavelength UV absorbance (UV254) was measured using a U-2010 instrument (Hitachi, Ibaraki, Japan). Prior to measuring DOC and UV254, the raw-river samples were filtered through Whatman glass microfiber filters (grade GF/F, 0.7-μm pore size; GE Healthcare, Buckinghamshire, United Kingdom) to remove suspended fractions, and other samples were directly analyzed.
Evaluation of the molecular-mass distribution of organic fractions by high-performance gel chromatography–size exclusion chromatography.
The molecular-mass distribution of organics in the samples was determined by using high-performance gel chromatography–size exclusion chromatography (HPGC-SEC) with a Protein Pak 125 column (Waters, Tokyo, Japan) that incorporated a UV254 detector (SPD-M10A VP Diode Array Detector, Shimadzu). Phosphate-buffered saline containing 1.39 g/liter of Na2HPO4, 1.39 g/liter of KH2PO4, and 3.51 g/liter of NaCl (pH 6.8) was used as the mobile phase (22). To obtain a calibration curve, 8.0 to 100 kDa of sodium polystyrene sulfonate was used.
Fractionation of organics by fluorescent excitation-emission matrix spectrophotometry.
To fractionate the organic contents in the samples, fluorescence EEM spectra were measured with an F-4500 fluorescence spectrophotometer (Hitachi). Excitation spectra between 200 nm and 400 nm (5-nm slit) and emission spectra between 230 nm and 600 nm (5-nm slit) were scanned. The measured values were calibrated by subtracting those obtained from a blank sample (Milli-Q water). Further, the fluorescence intensity of 10-μg/liter quinine sulfate solution (in 0.1 M H2SO4) at a 345-nm excitation wavelength/450-nm emission wavelength was considered 10 arbitrary units (AU) for the relative fluorescence intensity calculations of each sample.
RESULTS
Virus detection efficiencies.
To evaluate the virus detection efficiency, PV was spiked into each water sample to a final concentration of 9.0 × 104 GC/μl and quantified by RT-qPCR. The levels of indigenous EV in the river water samples were determined to be 1.9 × 100 GC/μl (at most), which were far below the spiked level, and therefore, the background EV levels were considered negligible. The virus detection efficiencies determined are summarized in Table 1. For the undiluted RNA extracts of humic acid solutions, the raw-Wa-HA, raw-Ald-HA, conc-Wa-HA, and conc-Ald-HA samples resulted in extremely low detection efficiencies (i.e., not detected to 0.1%), whereas other samples showed respectable values (i.e., 67 to 160%). Among the river water samples, all the raw-river-water samples showed high efficiencies (i.e., 89 to 140%), while all the conc-river-water samples showed low efficiencies (i.e., not detected to 9.9%).
TABLE 1.
Geometric mean recoveries and geometric standard deviations of PV and PV RNA spiked into samples determined by RT-qPCR
| Sample | Sample type | Spiked PV viriona |
Spiked PV RNAb (nondilution) |
||||
|---|---|---|---|---|---|---|---|
| Nondilution |
100-fold dilution |
||||||
| GMc (%) | GSDc | GM (%) | GSD | GM (%) | GSD | ||
| Blank (Milli-Q) | Raw | 100 | 0.08 | ||||
| Filt | 67 | 0.04 | |||||
| Acid | 70 | 0.05 | |||||
| Conc | 100 | 0.03 | |||||
| Wa-HA | Raw | 0.030 | 0.14 | 56 | 0.05 | ||
| Filt | 91 | 0.18 | |||||
| Acid | 106 | 0.08 | |||||
| Conc | 0.10 | 0.39 | 33 | 0.44 | |||
| Ald-HA | Raw | <0.0001d | 53 | 0.28 | |||
| Filt | 110 | 0.17 | |||||
| Acid | 130 | 0.26 | |||||
| Conc | 0.00078 | 0.14 | 73 | 0.05 | |||
| IH-HA | Raw | 130 | 0.26 | <0.0001 | - | ||
| Filt | 150 | 0.19 | 5.2 | 0.22 | |||
| Acid | 160 | 0.14 | |||||
| Conc | 130 | 0.24 | |||||
| R1 | Raw | 89 | |||||
| Conc | 9.9 | 62 | |||||
| R2 | Raw | 140 | |||||
| Conc | <0.0001 | 150 | |||||
| R3 | Raw | 110 | |||||
| Conc | <0.0001 | 77 | |||||
PV virions were spiked into the samples just before the RNA extraction step.
Extracted PV RNA was spiked into the samples to a final concentration of 4.5 × 104 GC/μl and quantified without an RNA extraction step.
GM and GSD, geometric mean and geometric standard deviation, respectively (n = 3 for blank and humic acid samples; n = 1 for river water samples; GSD for river water samples cannot be shown).
<0.0001% indicates that spiked PV or PV RNA was not detected by RT-qPCR.
Next, the RNA extracts that showed substantially low PV detection efficiencies were diluted 100-fold, and the RT-qPCR assay was repeated with the diluted samples. After the dilution, the detection efficiencies were increased for all the diluted samples (Table 1). Because the efficiencies were increased by diluting the RNA extracts, the extremely low PV detection efficiencies were presumably due to RT-qPCR inhibition. Hence, it can be concluded that the raw-HA and conc-HA samples of Wa-HA and Ald-HA and the conc-river-water samples contained sufficient amounts of RT-qPCR inhibitors to hinder PV detection by RT-qPCR. In addition, it also indicates that most of the inhibitors in Wa-HA and Ald-HA remained on the electronegative membrane until the elution step of sample concentration.
It was also presumed that IH-HA contains RT-qPCR inhibitors; however, all IH-HA samples showed high PV detection efficiency. To test the hypothesis that RT-qPCR inhibitors contained in the IH-HA samples are removed in the RNA extraction step, the raw-IH-HA and filt-IH-HA samples were spiked with extracted PV RNA originating from the purified PV (9.0 × 106 GC/μl) to a final concentration of 4.5 × 104 GC/μl, and the RT-qPCR assay was performed without the RNA extraction step. The results showed “not detected” for the raw-IH-HA sample and 5.2% detection efficiency for the filt-IH-HA samples. The results indicate that IH-HA contained an inhibitor that can be removed in the RNA extraction step. Considerable amounts of the inhibitors probably passed through the membrane during the first filtration step (adsorption step).
DOC and UV254 in the samples.
Figure 2 summarizes the DOC and UV254 results for the raw-HA samples and the loss or recovery ratio (a ratio comparing the values in the raw-HA sample and corresponding filt-HA, acid-HA, and conc-HA samples). For Wa-HA and Ald-HA, the DOC recovery ratios for the conc-HA samples were greater than 60% and the loss ratios for the filt-HA and acid-HA samples were ∼20% and <0.1%, respectively. Their UV254 recovery/loss ratios also showed similar tendencies. On the other hand, the DOC and UV254 loss ratios in the filt-IH-HA sample were 67% and 69%, respectively, and the loss ratio for the acid-IH-HA sample and the recovery ratios for the conc-HA sample were below 6.3%.
FIG 2.
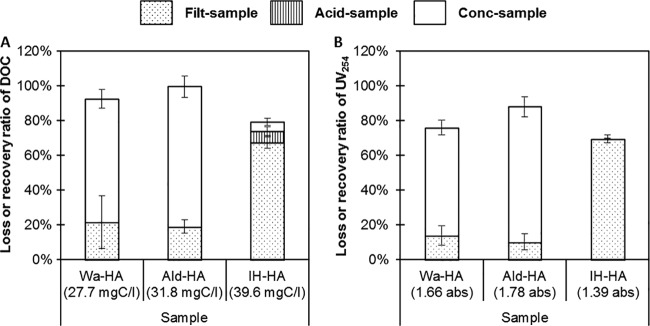
Loss or recovery efficiencies of DOC (A) and UV254 (B) in raw samples of humic acid solutions during the virus concentration process. The values indicate the concentrations in each raw sample. The error bars indicate standard deviations (n = 3).
Table 2 compares the DOC concentrations and UV254 responses for the raw-river-water and conc-river-water samples. The DOC concentrations and UV254 responses of the conc-river-water samples were 2.4 to 3.9 times higher than those of the raw-river-water samples. Note that these values were substantially lower than the virus concentration ratio (150-fold; 30 liters of raw samples were concentrated to 0.2 liters). It appears that some organics in the original sample are less concentrated than viruses in the final samples. It appears that the some organic substances are selectively recovered in the conc-river-water samples, as shown in the case of the humic acid solutions (Fig. 2).
TABLE 2.
DOC and UV254 in raw and concentrated samples of river water
| Sample | DOC |
UV254 (absorbance) |
||||
|---|---|---|---|---|---|---|
| Raw (mgC/liter) | Conc (mgC/liter) | Conc/raw | Raw (mgC/liter) | Conc (mgC/liter) | Conc/raw | |
| R1 | 1.2 | 3.5 | 2.9 | 0.035 | 0.120 | 3.4 |
| R2 | 1.2 | 4.8 | 3.9 | 0.025 | 0.060 | 2.4 |
| R3 | 1.0 | 2.5 | 2.6 | 0.027 | 0.088 | 3.3 |
Molecular mass distributions of organics in the samples.
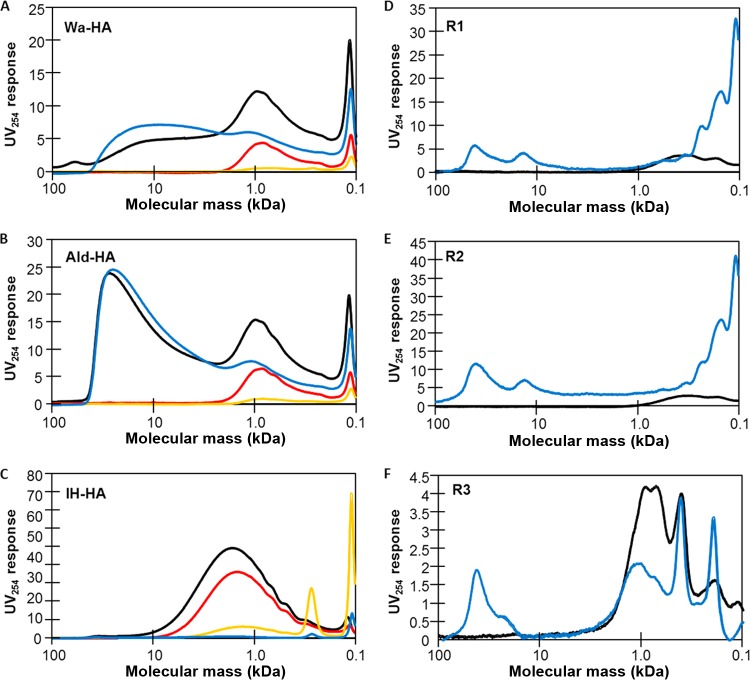
Because the results indicated that some organics are selectively recovered with the virus concentrate, the organics that are recovered and that may cause RT-qPCR inhibition were characterized based on their molecular masses. The molecular mass distribution of organics present in the samples was determined by HPGC-SEC. As shown in Fig. 3A, the raw-Wa-HA sample produced a moderate UV response peak between 10 and 100 kDa and a large peak around 1.0 kDa (peak intensity, 12.3 AU). The conc-Wa-HA sample also exhibited a peak between 10 and 100 kDa, which is approximately that of the raw-Wa-HA sample. On the other hand, the filt-Wa-HA and acid-Wa-HA samples developed no peaks in the region between 10 and 100 kDa. These results suggest that the organics with 10- to 100-kDa masses existing in the filt-Wa-HA and acid-Wa-HA samples were retained on the electronegative membrane during the sample concentration process and were efficiently recovered in the final sample concentrate. The filt-Wa-HA and conc-Wa-HA samples developed a similar UV response peak around 1.0 kDa. The sum of their peak intensities (6.6 AU and 7.9 AU in filt-Wa-HA and conc-Wa-HA, respectively) is comparable to that of the raw-Wa-HA sample, indicating that a part of the organic fraction passed through the electronegative membrane during the filtration process while the remaining part was recovered in the final sample concentrate. Similar trends were seen with the samples of Ald-HA (Fig. 3B). The raw-IH-HA sample showed a peak around 1.0 kDa (peak intensity, 43.6 AU) but exhibited no peaks in the 10- to 100-kDa range. The filt-IH-HA sample also showed a peak around 1.0 kDa with an intensity of 32.4 AU, which is similar to that observed with the raw-IH-HA sample. No notable peaks are shown in the region of >1.0 kDa for the acid-IH-HA and the conc-IH-HA samples (Fig. 3C). These results indicate that most of the organic fraction passed through the electronegative membrane during the filtration process.
FIG 3.
Molecular mass distribution of organics in Wa-HA (A), Ald-HA (B), IH-HA (C), R1 (D), R2 (E), and R3 (F) determined by UV254-based HPGC-SEC. The black, red, orange, and blue lines correspond to raw, filt, acid, and conc samples, respectively.
The raw-river-water and conc-river-water samples were also analyzed by HPGC-SEC (Fig. 3D to F). All of the conc-river-water samples showed peaks in the 10- to 100-kDa range, while none of the raw-river-water samples showed notable peaks in the same range. The raw-R3 sample showed notable peaks around 1.0 kDa (peak intensities: around 4.2 AU), which were also seen in the conc-R3 sample with lower peak intensities (∼1.9 AU).
EEM.
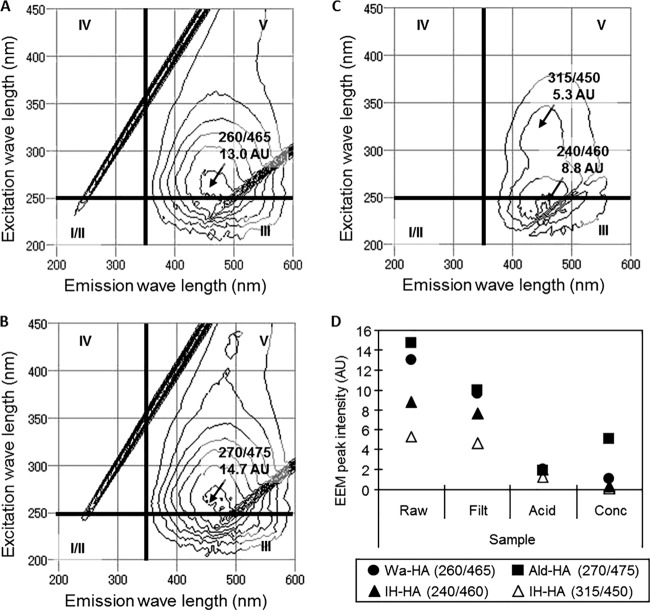
For further characterization of the organics present in the samples, EEM was performed. Figure 4A to C shows EEM spectra of the raw-Wa-HA, raw-Ald-HA, and raw-IH-HA samples, respectively. For the purpose of interpretation, each EEM spectrum is divided into five regions (regions I to V). Generally, the peaks appearing in regions I, II, and IV indicate the presence of protein-like compounds, and those appearing in regions III and V indicate the presence of fulvic acid-like and humic acid-like compounds, respectively (20). Because the raw-HA samples exhibited peaks in region V, these samples likely contained humic acid-like compounds (Fig. 4A to C). The intensity variations of the peaks during the virus concentration process are summarized in Fig. 4D. As can be seen, the highest EEM peak intensity occurred with the raw-HA samples and a lower intensity with the conc-HA samples. The results indicate that the humic acid-like compounds were not efficiently recovered in the virus concentrates.
FIG 4.
(A to C) EEM spectra in raw samples of Wa-HA (A), Ald-HA (B), and IH-HA (C). (D) Variation of intensities of the peaks seen in the raw samples during the virus concentration process. The values indicate the positions and intensities of the peaks. The EEM spectra are divided into 4 regions, I/II, III, IV, and V. The peaks appearing in regions I/II and IV correspond to protein-like substances, and those in regions III and V correspond to fulvic acid-like and humic acid-like substances, respectively.
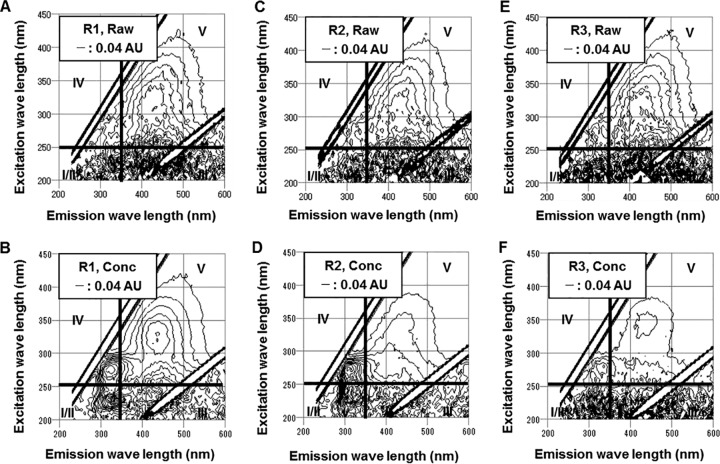
The raw-river-water and conc-river-water samples were also subjected to the EEM assay (Fig. 5). The raw-river-water samples showed peaks in region V (humic acid-like) with a peak intensity around 0.20 AU. The conc-river-water samples showed peaks in regions I/II, IV (both protein-like), and V. The intensities of the peaks that appeared in region V were 0.08 to 0.20 AU, which are similar to or lower than those of the raw-river-water samples. Considering that the volume was reduced in the virus concentration process (150-fold reduction), it seems that the compounds that generated the peak in region V were poorly recovered in the virus concentrate. The peaks in regions I/II and IV, which correspond to protein-like substances, were not observed in the raw-river-water samples, indicating that the protein-like substances were efficiently recovered in the virus concentrate.
FIG 5.
EEM spectra in raw samples of R1 (A), R2 (C), and R3 (E) and conc samples of R1 (B), R2 (D), and R3 (F).
DISCUSSION
Accurate quantification of enteric viruses in the water environment is important for public health. Although sensitivity is one of the strengths of RT-qPCR, it becomes unreliable in the presence of humic acid and other natural organics, which tend to be recovered by virus concentration methods (8, 10, 25). In this study, the organic properties (i.e., quantity, molecular mass distribution, and EEM spectra) of the water samples containing commercially available humic acids and river water samples were analyzed before, during, and after the concentration step.
Humic acid is defined as organic fractions that adsorb onto and elute from hydrophobic resin under low- and high-pH conditions, respectively, and that precipitate under low-pH conditions (19). Humic acid fractions extracted by the IHSS method have been characterized using their molecular masses of around 1.0 kDa and generate a peak in region V of their EEM spectra (21). In the present study, Wa-HA and Ald-HA possessed similar organic properties. Specifically, these HA samples after the virus concentration step showed a high recovery of organics and led to RT-qPCR inhibition. Furthermore, the samples contained organic fractions in the range of 10 to 100 kDa, which were efficiently recovered in the virus concentrates. On the other hand, most of the organics present in the IH-HA samples were lost during the virus concentration step. The RT-qPCR inhibitors contained in IH-HA were excluded during the RNA extraction step. The HPGC-SEC results revealed that IH-HA contained organic fractions of around 1.0 kDa, which was also observed with Wa-HA and Ald-HA. However, IH-HA did not contain organic fractions at 10 to 100 kDa. Comparison of the Wa-HA, Ald-HA, and IH-HA results suggests that the organic fractions of 10 to 100 kDa, which were recovered in the concentrates and RNA extracts, are the probable cause of the RT-qPCR inhibition. In the EEM analysis, these three humic acid samples showed peaks that indicate the presence of humic acid-like compounds. The HPGC-SEC results show that the behavior of the humic acid-like compounds exhibited in the virus concentration step was similar to that of the organic fraction of around 1.0 kDa (Fig. 3 and 4D). Considering the characteristics of humic acid extracted and purified by the IHSS method, the humic acid-like peaks seen in the EEM analysis and the organic fraction of around 1.0 kDa found in the HPGC-SEC analysis likely originated from humic acids that existed in the original samples. The organic fractions of 10 to 100 kDa in Wa-HA and Ald-HA appeared to be RT-qPCR inhibitors, but no corresponding EEM peak was observed. Considering their molecular masses and the absence of EEM peaks, these fractions can be identified as substances that possess some characteristics common to humic acids.
The river water concentrates caused RT-qPCR inhibition and contained organic fractions of 10 to 100 kDa, which were not seen in their original samples. Using a DOC detector for the SEC analysis, Kawasaki et al. (22) observed organic fractions of 10 to 100 kDa in lake water samples, but they did not observe these fractions in the UV254-based SEC analysis. This indicates that 10- to 100-kDa organic fractions may be present in environmental water even though the concentrations of the fractions are too low to be detected by UV254-based SEC. In this study, 30 liters of river water samples was concentrated to 0.2 liters by the adsorption-elution method (150-fold concentration). Considering the concentration ratio, it is possible that the organic fractions of 10 to 100 kDa were present in the raw-river-water samples at undetectable levels and that they were efficiently concentrated to a detectable level. The 10- to 100-kDa fractions were also observed in the raw-Wa-HA/raw-Ald-HA and conc-Wa-HA/conc-Ald-HA samples and were assumed to be RT-qPCR inhibitors. It is likely that the 10- to 100-kDa organic fractions observed in the river water samples acted as RT-qPCR inhibitors. In addition to the 10- to 100-kDa organic fractions, those around 1.0 kDa were also observed in the conc-R3 sample. Similar fractions were also seen in conc-Wa-HA/conc-Ald-HA samples but were not seen in the conc-IH-HA sample. Considering that the river water and Wa-HA/Ald-HA samples contained 10- to 100-kDa organic fractions, it is possible that the 10- to 100-kDa organic fractions mediated the recovery of the 1.0-kDa organic fractions recovered in the concentrate. However, as the peak intensities of the 1.0-kDa organic fractions in the conc-R3 sample were lower than those in the original sample, which did not result in RT-qPCR inhibition, it is unlikely that the fractions were RT-qPCR inhibitors. The EEM results revealed that protein-like substances were well recovered in the concentrates. Because the surface constituents of nonenveloped virus particles are proteins, it is possible that the concentration method used in this study can recover proteins other than virus particles. In the RNA extraction step, a guanidine-based proteinase was used to remove proteins; thus, the protein-like substances were unlikely to cause RT-qPCR inhibition. This can be clarified if the RNA extracts are subjected to organic analysis. However, the volume of the RNA extract (60 μl) was too small for our analysis, which requires ∼10 ml of a sample. Diluting the RNA extract can enlarge the sample volume but may decrease the organics below the detectable level. A possible future improvement would be increasing the concentration ratio to recover higher concentrations of the organics.
All humic acid and river water samples that resulted in RT-qPCR inhibition contained organic fractions of 10 to 100 kDa, and they did not seem to generate any EEM peaks. These fractions were efficiently recovered in the sample concentrates and very likely led to RT-qPCR inhibition. In previous studies (7, 9, 12), an ultrafiltration (UF) membrane with a molecular mass cutoff of 30 to 50 kDa was used as a secondary concentration method to detect viruses in environmental samples. The high-molecular-mass fractions observed in this study were probably what remained in the UF concentrate. Because the 10- to 100-kDa fractions were recovered by the sample concentration method using an electronegative membrane, these organics were possibly trapped or adsorbed onto the membrane in the presence of Mg2+ in the sample. The fractions remained on the membrane during the acid rinse step and then dispersed during the alkaline elution step. In this study, precipitates were formed in the Wa-HA and Ald-HA samples, which were most likely trapped on the membrane in the presence of Mg2+ under acidic conditions (data not shown).
It has been pointed out that organic inhibitors such as humic acid affect the RT-PCR-based detection of viruses in water (8, 10, 25). However, few studies have tried to identify the inhibitors in field samples, and at present, there is no standard method that gives a reliable detection efficiency (8, 15). Abbaszadegan et al. (25) compared DOC, UV254, and 254-nm specific UV absorbance in raw water samples to virus detection efficiencies, but they found only poor correlations between them. Our previous study also found no clear correlations between the organic indicators in raw water samples and the virus detection efficiencies (11). Our study indicates that some specific organic substances are selectively recovered in the virus concentrates and adversely influence virus detection efficiencies. Therefore, characterization of inhibitors by analyzing raw-water sample and final sample concentrates is required for future establishment of a method to improve virus detection efficiency. Rock et al. (26) applied EEM analysis to virus extracts obtained from biosolid samples using beef extract and glycine buffer and found peaks of humic acid-like and fulvic acid-like substances. However, the relationship between the virus detection efficiency and the EEM peaks was unclear. In this study, DOC, UV254, HPGC-SEC, and EEM were used to characterize organic fractions in samples during the virus concentration processes. Our results indicate that natural organic fractions in the 10- to 100-kDa size range adversely affect the efficiency of virus detection by RT-qPCR. Since the size of poliovirus, a typical enteric virus, is larger than 1,000 kDa (4), size exclusion sieving techniques such as gel chromatography may be effective for separation of enteric viruses from the inhibitors. However, as was suggested by a previous study (7), the variety of organics in environmental samples may affect RT-qPCR-based virus detection. The causes of inhibition may also be different depending on the virus concentration method applied. Thus, it is probable that the suitability of a sample purification technique depends on the samples and sample treatments. It is important to characterize the inhibitors in a variety of environmental samples by examining the relationships among dissolved organics in the original samples and their virus concentrates and virus detection efficiencies for the establishment of an effective method for sample clarification.
ACKNOWLEDGMENT
This work was supported by a grant-in-aid for JSPS Fellows (grant number 12J08885) from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Sinclair RG, Jones EL, Gerba CP. 2009. Viruses in recreational water-borne disease outbreaks: a review. J Appl Microbiol 107:1769–1780. doi: 10.1111/j.1365-2672.2009.04367.x. [DOI] [PubMed] [Google Scholar]
- 2.Villena C, Gabrieli R, Pintó RM, Guix S, Donia D, Buonomo E, Palombi L, Cenko F, Bino S, Bosch A, Divizia M. 2003. A large infantile gastroenteritis outbreak in Albania caused by multiple emerging rotavirus genotypes. Epidemiol Infect 131:1105–1110. doi: 10.1017/S0950268803001353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson KE, Schwab KJ, Spencer SK, Borchardt MA. 2012. Measuring and mitigating inhibition during quantitative real time PCR analysis of viral nucleic acid extracts from large-volume environmental water samples. Water Res 46:4281–4291. doi: 10.1016/j.watres.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 4.Ohlbaum A, Figueroa F, Grado C, Contreras G. 1970. Target molecular weight of foot-and-mouth disease virus and poliovirus. J Gen Virol 6:429–432. doi: 10.1099/0022-1317-6-3-429. [DOI] [PubMed] [Google Scholar]
- 5.Ye XY, Ming X, Zhang YL, Xiao WQ, Huang XN, Cao YG, Gu KD. 2012. Real-time PCR detection of enteric viruses in source water and treated drinking water in Wuhan, China. Curr Microbiol 65:244–253. doi: 10.1007/s00284-012-0152-1. [DOI] [PubMed] [Google Scholar]
- 6.da Silva AK, Le Saux J-C, Parnaudeau S, Pommepuy M, Elimelech M, Le Guyader FS. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl Environ Microbiol 73:7891–7897. doi: 10.1128/AEM.01428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hata A, Katayama H, Kitajima M, Visvanathan C, Nol C, Furumai H. 2011. Validation of internal controls for extraction and amplification of nucleic acids from enteric viruses in water samples. Appl Environ Microbiol 77:4336–4343. doi: 10.1128/AEM.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrader C, Schielke A, Ellerbroek L, Johne R. 2012. PCR inhibitors—occurrence, properties and removal. J Appl Microbiol 113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 9.Cashdollar JL, Wymer L. 2013. Methods for primary concentration of viruses from water samples: a review and meta-analysis of recent studies. J Appl Microbiol 115:1–11. doi: 10.1111/jam.12143. [DOI] [PubMed] [Google Scholar]
- 10.Wilson IG. 1997. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 63:3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hata A, Katayama H, Kojima K, Sano S, Kasuga I, Kitajima M, Furumai H. 2014. Effects of rainfall events on the occurrence and detection efficiency of viruses in river water impacted by combined sewer overflows. Sci Total Environ 468-469:757–763. doi: 10.1016/j.scitotenv.2013.08.093. [DOI] [PubMed] [Google Scholar]
- 12.Fernández MD, Torres C, Poma HR, Riviello-López G, Martínez LC, Cisterna DM, Rajal VB, Nates SV, Mbayed VA. 2012. Environmental surveillance of norovirus in Argentina revealed distinct viral diversity patterns, seasonality and spatio-temporal diffusion processes. Sci Total Environ 437:262–269. doi: 10.1016/j.scitotenv.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 13.Hamza IA, Jurzik L, Stang A, Sure K, Uberla K, Wilhelm M. 2009. Detection of human viruses in rivers of a densly-populated area in Germany using a virus adsorption elution method optimized for PCR analyses. Water Res 43:2657–2668. doi: 10.1016/j.watres.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Katayama H, Shimasaki A, Ohgaki S. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl Environ Microbiol 68:1033–1039. doi: 10.1128/AEM.68.3.1033-1039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory JB, Litaker RW, Noble RT. 2006. Rapid one-step quantitative reverse transcriptase PCR assay with competitive internal control for detection of enteroviruses in environmental samples. Appl Environ Microbiol 72:3960–3967. doi: 10.1128/AEM.02291-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreader CA. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol 62:1102–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodríguez RA, Thie L, Gibbons CD, Sobsey MD. 2012. Reducing the effects of environmental inhibition in quantitative real-time PCR detection of adenovirus and norovirus in recreational seawaters. J Virol Methods 181:43–50. doi: 10.1016/j.jviromet.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Schriewer A, Wehlmann A, Wuertz S. 2011. Improving qPCR efficiency in environmental samples by selective removal of humic acids with DAX-8. J Microbiol Methods 85:16–21. doi: 10.1016/j.mimet.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 19.International Humic Substances Society. 2007. What are humic substances? http://www.humicsubstances.org/whatarehs.html.
- 20.Chen W, Westerhoff P, Leenheer JA, Booksh K. 2003. Fluorescence excitation-emission matrix regional integration to quantify spectra for dissolved organic matter. Environ Sci Technol 37:5701–5710. doi: 10.1021/es034354c. [DOI] [PubMed] [Google Scholar]
- 21.Chin YP, Aiken G, O'Loughlin E. 1994. Molecular weight, polydispersity, and spectroscopic properties of aquatic humic substances. Environ Sci Technol 28:1853–1858. doi: 10.1021/es00060a015. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki N, Matsushige K, Komatsu K, Kohzu A, Nara FW, Ogishi F, Yahata M, Mikami H, Goto T, Imai A. 2011. Fast and precise method for HPLC-size exclusion chromatography with UV and TOC (NDIR) detection: importance of multiple detectors to evaluate the characteristics of dissolved organic matter. Water Res 45:6240–6248. doi: 10.1016/j.watres.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 23.Imai A, Fukushima T, Matsushige K, Kim YH, Choi K. 2002. Characterization of dissolved organic matter in effluents from wastewater treatment plants. Water Res 36:859–870. doi: 10.1016/S0043-1354(01)00283-4. [DOI] [PubMed] [Google Scholar]
- 24.Hata A, Matsumori K, Kitajima M, Katayama H. 18 October 2014. Concentration of enteric viruses in large volumes of water using a cartridge-type mixed cellulose ester membrane. Food Environ Virol doi: 10.1007/s12560-014-9169-x. [DOI] [PubMed] [Google Scholar]
- 25.Abbaszadegan M, Stewart P, LeChevallier M. 1999. A strategy for detection of viruses in groundwater by PCR. Appl Environ Microbiol 65:444–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rock C, Alum A, Abbaszadegan M. 2010. PCR inhibitor levels in concentrates of biosolid samples predicted by a new method based on excitation-emission matrix spectroscopy. Appl Environ Microbiol 76:8102–8109. doi: 10.1128/AEM.02339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]