Abstract
Volcanic eruptions are a widespread force of geological and ecological disturbance and present recurrent opportunities for the study of biological responses to novel habitat formation. However, scientific study of such events is difficult given their short duration and often distant location. Here we report results from opportunistic sampling of unique volcano-generated habitats formed during the 2011 explosive eruption in the Puyehue-Cordón Caulle complex (Chile), when massive amounts of pumice were ejected, creating novel floating substrata that have never before been characterized from a microbiological perspective. DNA sequencing revealed a dynamic community of microbes that came to inhabit the pumice, with a unique composition distinct from that of the lakes' surface waters and with suggestions of ecological convergence across lakes and sampling times. Furthermore, biogeochemical studies of net nutrient fluxes showed that, while the fresh pumice arriving to the lakes was an initial source of phosphorus (P), colonized pumice had high rates of nitrogen (N) and P uptake and was sufficiently abundant to represent a significant lake-wide nutrient sink. These findings highlight the remarkable versatility of microbes in exploiting novel environments and are consistent with a recent proposal of floating pumice as a favorable environment for the initial origins of life on early Earth.
INTRODUCTION
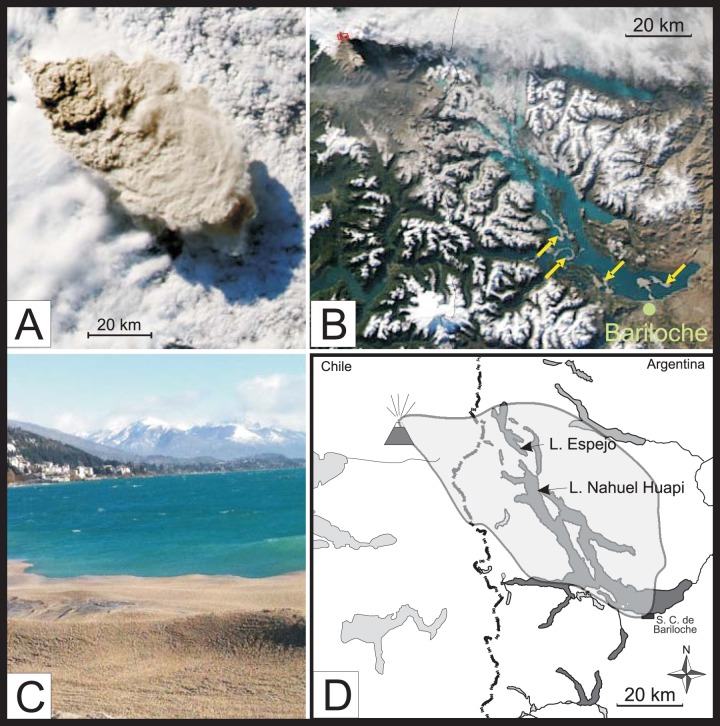
Volcanism is an active and powerful force shaping landscapes globally. Volcanic eruptions are of particular relevance not only because they are important in formation of local landforms but also because they impact more distant ecosystems by production and aerial transport of ash, pumice (a light and porous rock formed when a gas-rich froth of lava solidifies), and other ejecta. Indeed, such massive perturbations present unique opportunities for scientific discovery (1). The best-studied case of a volcanically impacted aquatic ecosystem is Spirit Lake (United States) following the Mt. St. Helens eruption (2). Investigations following that eruption resulted not only in a better understanding of volcanic impacts but also in discovery of novel biota that prospered posteruption, such as a new species of Legionella (3). On 4 June 2011 an explosive eruption began at the Puyehue-Cordón Caulle volcanic complex (40°31′24.28″S, 72°8′47.10″W in southern Chile), the first since 1960. This eruption (Fig. 1A) provided an opportunity to document ecological responses associated with volcanic eruption because, unlike the situation at Spirit Lake and other ecosystems impacted by an eruption, the lakes of this region were already under intensive study by local scientists (4–8). Early phases of the pyroclastic event produced plumes of ash that reached an altitude of 15 km and subsequently circled the globe. The eruption ejected large quantities of rhyolitic pumice (∼65 tons per hectare in areas close to the volcano [9]) that traveled as far as 100 km to the southeast (Fig. 1A). We earlier reported the effects of suspended ash on limnological conditions in the lakes impacted by Puyehue-Cordón Caulle (10). Here we focus not on the ecological impacts of the eruption in the lakes per se but instead on the emergence of unique and largely unstudied novel habitats that followed the eruption: large, island-like aggregations of floating pumice rocks (0.1 to 5 cm in diameter) (Fig. 1B and C). We focused on those in Lake Espejo and northwest Lake Nahuel Huapi (LNH) (Fig. 1D), which received 15 to 25 cm of direct aerial input of ash and pumice during the ∼8 h after the eruption (Fig. 1A). These rafts persisted for months and indeed, in lower abundance, for nearly 3 years following the eruption. Anecdotes of such pumice islands have been reported for the oceans, and indeed such formations have been considered potential vectors of biogeographic dispersal for marine and terrestrial biota (11, 12). Moreover, floating pumice has been proposed as a substrate for early chemical evolution and the origin of life because of its combination of chemically reactive surfaces and small interior voids that could protect early protobiological molecules and provide a stable environment for various reactions (13, 14). However, to our knowledge, floating pumice has never been characterized from a microbiological perspective using modern genetic techniques, nor have its biogeochemical impacts been examined quantitatively.
FIG 1.
(A) The initial plume of the Puyehue-Cordón Caulle eruption on 4 June 2011. Note that the direction of transport is to the southeast, bringing massive amounts of ash and pumice to Lake Nahuel Huapi (not visible under the plume and cloud cover). (NASA image courtesy of Jeff Schmaltz, MODIS Rapid Response Team, NASA/Goddard Space Flight Center.) (B) Two weeks after the initial eruption (20 June 2011), islands of pumice (gray formations, four of which are highlighted with yellow arrows) are visible in Lake Nahuel Huapi. (NASA image courtesy of Jeff Schmaltz, MODIS Rapid Response Team, NASA/Goddard Space Flight Center.) (C) A raft of pumice washes ashore at the southeast end of Lake Nahuel Huapi, near Bariloche. (Photo courtesy of E. Balseiro.) (D) Map showing the two main study lakes in relation to the plume depicted in panel A. (Adapted from reference 10.)
Here we report the results of sampling and experimental studies of the floating pumice originating from the Puyehue-Cordón Caulle volcanic eruption. During the ensuing austral summer in northern Lake Nahuel Huapi and in the smaller Lake Espejo (both ∼35 km from the eruption site), we assessed the microbial community structures associated with the pumice and with the accompanying lake water using bacterial 16S rRNA gene pyrosequencing, comparing the bacterial taxa found on the pumice with those in paired water samples. Our aims were to determine if the pumice developed a characteristic assemblage distinct from that found in the lake water and if pumice microbial communities at different times and in different lakes had similar or divergent structures. We also assessed the direct and indirect effects of pumice inputs by characterizing fluxes associated with fresh and colonized pumice, comparing these to the magnitude of in-lake nutrient fluxes.
MATERIALS AND METHODS
Study sites and field measurements.
Pumice sampling was carried out in two lakes, Lake Espejo and Lake Nahuel Huapi, that are part of the glacial lake district of the North Patagonian Andes (15). Lake Espejo and northwest Lake Nahuel Huapi (Angostura arm) are close to the eruption (∼35 km) and were subjected to considerable pumice input during the initial explosion. Indeed, after the eruption, both lakes were partially covered by large pumice rafts visible from satellite (Fig. 1B, arrows). Single-grab samples of pumice, along with ambient lake water, were collected opportunistically as the pumice floated at the surface at various locations. Pumice was obtained by scooping with sterile sample tubes; sterile technique was used. Water samples were obtained by submerging an acid-cleaned bottle (rinsed with lake water) under water and then allowing it to fill with surface water while preventing pumice from entering. As described below, pumice and water samples were held in a cooler until they could be processed for DNA extraction and other procedures (within 5 h generally).
During the initial eruption, free-falling pumice was collected within the Bariloche city limits by placing plastic buckets and zippered bags on the ground for 1 to 2 h during the night of the eruption (ca. 1600 to 2000 h on 4 June 2014). Pumice collected in this manner was then immediately transferred to zippered plastic bags and stored unopened until being used for experiments. Throughout the posteruption period (September 2011 to March 2012), pumice in the lakes was sampled opportunistically from a boat. For DNA extraction, pumice and water samples were obtained directly at the surface using sterilized 50-ml tubes. Samples were carried to the laboratory in thermally isolated containers within 3 h after sampling and were processed immediately after arrival to the laboratory. Pumice samples were gently washed with 0.2-μm-filtered lake water to remove all particles other than the rocks and their attached organisms. To obtain planktonic bacteria for sequencing, 100 ml of lake water was concentrated on 0.2-μm sterile hydrophilic polyether sulfone Supor filters (Gelman Laboratory, Inc.). Processed samples (pumice and filters) were flash frozen in liquid nitrogen and stored at −80°C until analysis. All materials used were presterilized.
To estimate the magnitude of pumice-associated nutrient fluxes in comparison to those in lake surface waters in the impacted region, we focused on Lake Espejo, for which we had relevant data on important parameters. We estimated the total amount of floating pumice present in the lake per unit area by assuming an areal input of pumice for Lake Espejo of ∼7.5 × 104 g m−2 based on reported ash and pumice deposition data for the region close to the lake (9). We also assumed that, of this deposition, 33% had a particle size of >1.5 mm. The actual particle size of deposited materials (e.g., the mix of fine ash and “lapilli” capable of floating) varied considerably with distance from the volcano and with time after the initial eruption; a value of 33% is within the range reported for materials collected with 40 to 60 km of the eruption during its immediate aftermath (16). Based on our own observations of lake surfaces during midsummer in comparison to the weeks immediately after the eruption, we also assumed that by midsummer about ∼1 to 10% of this material remained floating in the lake. This yielded areal estimates of midsummer pumice abundance in heavily impacted lakes such as Lake Espejo and northwestern Lake Nahuel Huapi ranging from 250 g m−2 to 2,500 g m−2.
DNA extraction and sequencing.
DNA was extracted from 10 g pumice using the MoBio PowerMax soil DNA isolation kit (MoBio Laboratories, Carlsbad, CA) following the manufacturer's protocol except for one modification (the cell disruption step was performed using a sonicator instead of a vortex). The pumice and bead mixture was sonicated using a Branson 450 cuphorn sonifier at duty cycle 50% and output 4. The sample was sonicated for four cycles of 2.5 min while resting on ice between each cycle. DNA from water samples was extracted using the MoBio PowerWater DNA isolation kit.
The 16S rRNA gene was amplified using barcoded primers targeting the V3 and V5 region of the gene (357F, 5′-CCTACGGGAGGCAGCAG; 926R, 5′-CCGTCAATTCMTTTRAGT-3′). Each 20-μl PCR mixture consisted of 0.3 μl Platinum Taq high-fidelity DNA polymerase (Life Technologies, Grand Island, NY), 1× high-fidelity PCR buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 2 mM MgSO4, 0.2 mM each primer, and 2 μl template DNA. The PCR conditions were as follows: denaturation at 95°C for 2 min, 30 cycles of 95°C for 20 s, 56°C for 30 s, and 72°C for 5 min, and final elongation at 72°C for 5 min. The amplicons were analyzed by gel electrophoresis and cleaned using the solid-phase reversible immobilization (SPRI) cleanup method with the Agencourt AMPure XP system (Beckman Coulter, Brea, CA) at a DNA-to-bead ratio of 1:5. The purified amplicons were quantified using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA) and mixed in equimolar concentration. The pooled DNA sample was shipped on dry ice to the University of Illinois W. M. Keck Center for Comparative and Functional Genomics for unidirectional multiplex pyrosequencing on the Roche GS FLX+ system.
Sequence analyses.
For the 10 samples, a total of 70,683 sequences with a median read length of 615 bp were obtained. The 16S rRNA sequences were screened for length (minimum of 100 bp), quality, and hexapolymeric and chimeric sequences using the bioinformatics program Mothur (17). In addition, the initial sequence data set was aligned at default settings with Ribosomal Database Project (RDP release 10.32) bacterial 16S rRNA aligned sequences as a template (downloadable at https://rdp.cme.msu.edu/misc/resources.jsp) and clustered (furthest neighbor) at a cutoff of 97% sequence identity. This initial clustering step was used to identify and remove operational taxonomic units (OTUs) from the data set with a representative sequence corresponding to chloroplasts in the RDP database, leaving 69,363 (98.1%) total sequences. Using Mothur, the remaining sequences were subsampled to the smallest group (1,609 sequences) for normalization across samples, yielding a total of 16,090 sequences for the 10 samples. The 16,090 subsampled sequences were realigned and reclustered into OTUs, with a 97% sequence identity cutoff and a representative sequence being chosen for each OTU.
Subsampled sequences were also clustered with Mothur at cutoffs from 0.60 to 0.99 sequence identity to explore the community structure at various taxonomic levels. The resulting clusters were used to create sequence alignment-oriented radial heat maps using Clustal Omega for multiple-sequence alignment and the Perl-based software package Circos (18, 19). Dendrograms depicting the relationships between individual samples of pumice/water pairs were constructed, with branch lengths calculated using Bray-Curtis similarity.
Taxonomy-level histograms were created from the OTUs in the subsampled data set. Analysis of similarity (ANOSIM) and similarity percentage (SIMPER) analyses, using Bray-Curtis dissimilarity, were used to test for a difference in OTU profiles between pumice and water groups and to determine the top OTU contributors to the dissimilarity (20). Representative sequences from each OTU were chosen based on minimum sequence identity distance to all other sequences in the OTU and then classified using the RDP classifier (21). Abundances for each taxa were summed by sample based on the taxonomic classification for each OTU. The “top” taxa were then chosen by highest total SIMPER contribution. Pumice/water skew values were calculated as skew = log[(P +1)/(W + 1)], where P = mean pumice sample taxon abundance and W = mean water sample taxon abundance.
Biogeochemical fluxes associated with pumice.
We estimated the potential release/uptake of N and P by the initial deposition of pumice via experiments using pumice collected directly from the air during the night of the 4 June 2011 eruption. To estimate the initial impact of this aerially delivered pumice on N and P availability, we measured net changes in NH3/4 and PO4 after ∼3- to 4-g replicates of this pumice were suspended in 100 ml of 0.2-μm-filtered water from Lake Nahuel Huapi and then incubated for 20 h at a temperature of 18°C in 150-ml sealed flasks on a shaker table (90 rpm). Changes were measured both for unenriched water and for water enriched to 8 μM NH3/4 and 0.5 μM PO4 (n = 3 for each treatment). Net changes in lake water concentrations were assessed via colorimetric analyses (22).
To evaluate biological uptake of nutrients by pumice microbes, nutrient uptake kinetics experiments using colonized pumice freshly collected for Lake Espejo were performed on 15 February 2012 at ambient lake temperature (18°C) with moderate illumination (64 μE m−2 s−1) and shaking (to simulate wave action on the lake surface). In these experiments, lake water was spiked with NH4, NO3, or PO4 to achieve six concentrations of each (from 0 to 36 μM N as NH4 and NO3 and from 0 to 4.5 μM P as PO4). Net changes in N and P concentrations after 2.5 h were assessed via colorimetric analyses (22). We then estimated the uptake kinetics for N and P by fitting the observed data to a Michaelis-Menten function to estimate the half-saturation constant (K1/2) and maximum uptake rate (Vmax). Vmax estimates were normalized to pumice mass.
For comparison of pumice nutrient uptake rates to microplankton nutrient demand in the mixed layer of Lake Espejo, we used experimental (10) and monitoring data from mid-January 2012 (specific growth rate of 0.1 day−1 and overall seston biomass in the upper mixed layer [4-m depth] of ∼3.2 × 104 μmol C m−2 with C/N/P ratios of 135:9:1 [molar]) (10). Multiplying the specific growth rate by C biomass per unit area and then this value by the observed seston N/C and P/C ratios yielded rates of microplankton N and P demand of 24 μmol P m−2 day−1 and 210 μmol N m−2 day−1.
Nucleotide sequence accession number.
All sequences were deposited in GenBank under accession number PRJNA254988.
RESULTS
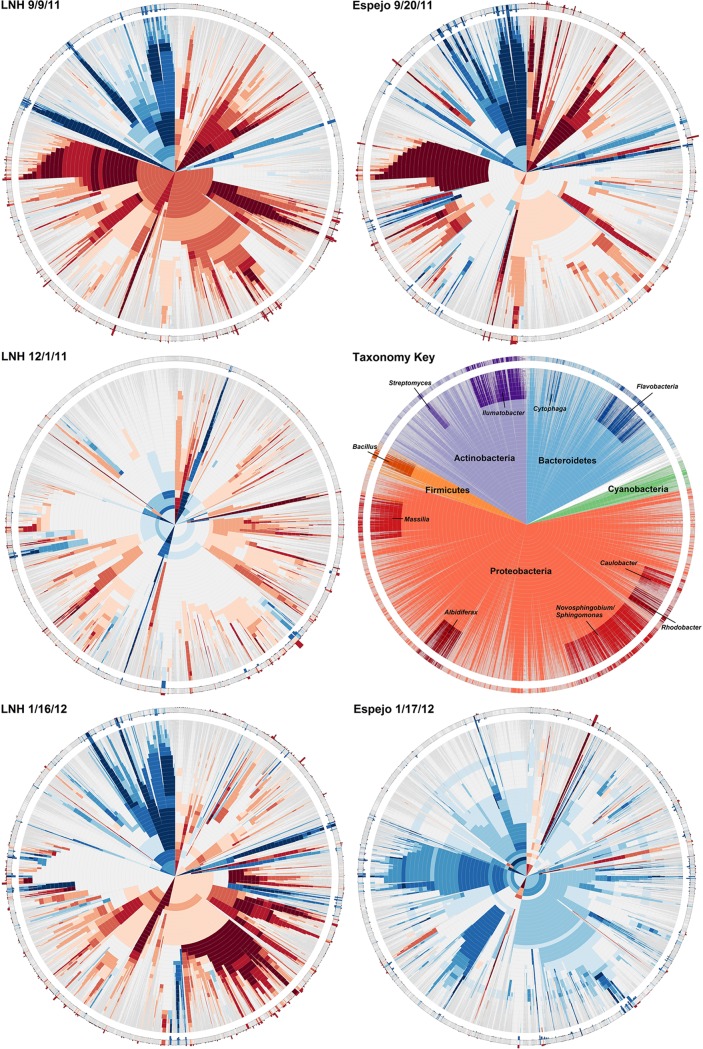
The 16S rRNA gene data set that emerged from our sampling comprised over 60,000 total individual reads (Table 1; see Table S1 in the supplemental material) that, as described in Materials and Methods, were subsampled to yield a total of 16,090 for analysis. Among these, members of the Proteobacteria dominated both the pelagic and the pumice communities (44% and 68%, respectively). However, pelagic communities included a secondary predominance of Actinobacteria (31%), which were relatively minor among the pumice microbes (∼3%), while members of the Bacteroidetes were common in pumice communities (26%) but less important in the water column (∼10%). A distinct nature of the pumice versus water communities was suggested by the community similarity analyses, which grouped three of the five water communities (LNH 9/9/11 and 1/16/12 and Espejo 9/20/11) together in a branch, while four of the five pumice communities (LNH 9/9/11, 12/1/11, 1/16/12, and Espejo 9/20/11) were grouped together in another branch (Fig. 2A). To more explicitly compare pumice and water communities, for each sample we analyzed relative abundances (as pumice/water ratios) at various levels of taxonomic similarity from 97% (species) to 66% (phyla) (Fig. 3). This analysis revealed two important patterns. First, the pumice-versus-water disparity is deeply rooted, as illustrated by the orange (pumice-loving) versus blue (water-loving) bands that penetrate to the center of Fig. 3. Second, the community structure of pumice-affiliated microbes was notably similar across lakes and dates for two closely nested pairs of samples obtained from different lakes and at different times (e.g., Lake Nahuel Huapi on 16 January 2012 nested with Lake Espejo on 20 September 2011, and Lake Nahuel Huapi on 1 December 2012 nested with Lake Espejo on 17 January 2012) (Fig. 2B).
TABLE 1.
Diversity indices and phylotype summary of water and pumice microbial communities
| Parameter | Valuea for: |
|
|---|---|---|
| Water | Pumice | |
| Observed OTUs | 311 (254, 375) | 336 (275, 381) |
| Chao richness estimator | 423 (294, 553) | 471 (340, 578) |
| Shannon diversity index | 5.30 (5.10, 5.62) | 5.51 (5.34, 5.72) |
| Shannon evenness | 0.658 (0.612, 0.745) | 0.742 (0.691, 0.803) |
| Simpson index of diversity | 0.991 (0.989, 0.994) | 0.993 (0.993, 0.995) |
| Top 4 dominant taxa by phylum (%) | Proteobacteria (44), Actinobacteria (31), Firmicutes (11), Bacteroidetes (10) | Proteobacteria (68), Bacteroidetes (26), Actinobacteria (3), Firmicutes (0.7) |
Values are presented as average (low, high) for the five samples of each type (water or pumice). All indices were calculated from a subsampled library that consisted of 16,090 reads total across the 10 samples.
FIG 2.

(A) Bray-Curtis similarity tree comparing the microbial communities in the 10 samples. Branch lengths were determined using the Bray-Curtis similarity index, which incorporates OTU presence and abundance for each sample (tree node). (B) Dendrogram of Bray-Curtis similarity tree comparing the relative pumice/water column abundance ratio for all OTUs identified in the samples. The ratio of all OTUs for a specific date is used as an overall measure of habitat preference. Branch lengths were determined using the Bray-Curtis similarity index, which incorporates OTU presence and abundance for each sample (tree node).
FIG 3.
Cladographic radial heat maps depicting the pumice/water skew of sequence clusters with a sequence divergence cutoff ranging from 0.4 to 0.03. Positioning of clusters on each graph is identical, arranged by multiple alignment of their containing sequences; thus, specific sections can be directly compared between graphs. Rings consist of sequence clusters at given cutoffs, starting from 0.4 in the center out to 0.03. Each cluster is then colored to represent its skew toward pumice-skewed sequences (red) or water-skewed sequences (blue). The outermost ring (cutoff, 0.03; equivalent to OTU) displays small histograms on its borders, representing the sequence abundance for pumice (red bars) and water (blue bars). A single sequence can be followed from the center of the graph and out to the edge in a straight line that has equal positioning in all graphs.
To highlight the microbial taxa most specialized to proliferate on these unique pumice habitats, we identified the most strongly pumice-affiliated taxa as well as the most pumice-avoiding (water-affiliated) taxa based on pumice/water ratio of observed counts at various phylogenetic levels (Fig. 4) for an aggregated data set involving all five sample pairs. Most phyla (12 out of 18) were underrepresented on pumice relative to the water, although one phylum (Thermotogae) was observed only on pumice and not in the water column (although these involved only a few reads) (Fig. 4, top panel). Notably, cyanobacteria were strongly underrepresented on the pumice (Fig. 4, top panel), a pattern that held true also for eukaryotic photoautotrophs as shown by 16S rRNA gene sequences representative of chloroplasts (data not shown), for which 1,311 reads (99.3% of the total across all samples, classified as cryptomonads, diatoms, and green algae) were found in water samples but only nine such reads (0.7%) were detected on pumice. At the level of order, strong skewing in favor of, and against, pumice colonization was observed (Fig. 4, middle panel). Indeed, 8 of the total 84 bacterial orders observed in the water column at least five times were not detected in the pumice samples. The order most underrepresented on pumice relative to the water column was family II, cyanobacteria (data not shown), consistent with the low representation of photoautotrophs on pumice observed at the phylum level and with the previously documented abundance of members of this group (e.g., Synechococcus) in the water columns of these lakes (23). Analysis at the genus level revealed strong habitat preferences consistent with the higher taxonomic levels (Fig. 4, bottom panel). For example, there were 35 genera that were detected at least five times in the water column but never on the pumice. This includes Bacillus, which was relatively abundant in Lake Nahuel Huapi on two dates. However, strongly pumice-affiliated taxa were also identified; indeed, of the 10 taxa that contributed most strongly to community-level patterns that distinguish pumice from water assemblages, six were skewed in favor of pumice. Notable among these are the highly disproportionate contributions of Cytophaga (∼10-fold skew), Caulobacter (∼6-fold skew), and Flavobacterium (∼3-fold skew) on pumice. Also noteworthy is strongly pumice-skewed Albidiferax (Proteobacteria), of which there is only one described species (A. ferrireducens).
FIG 4.
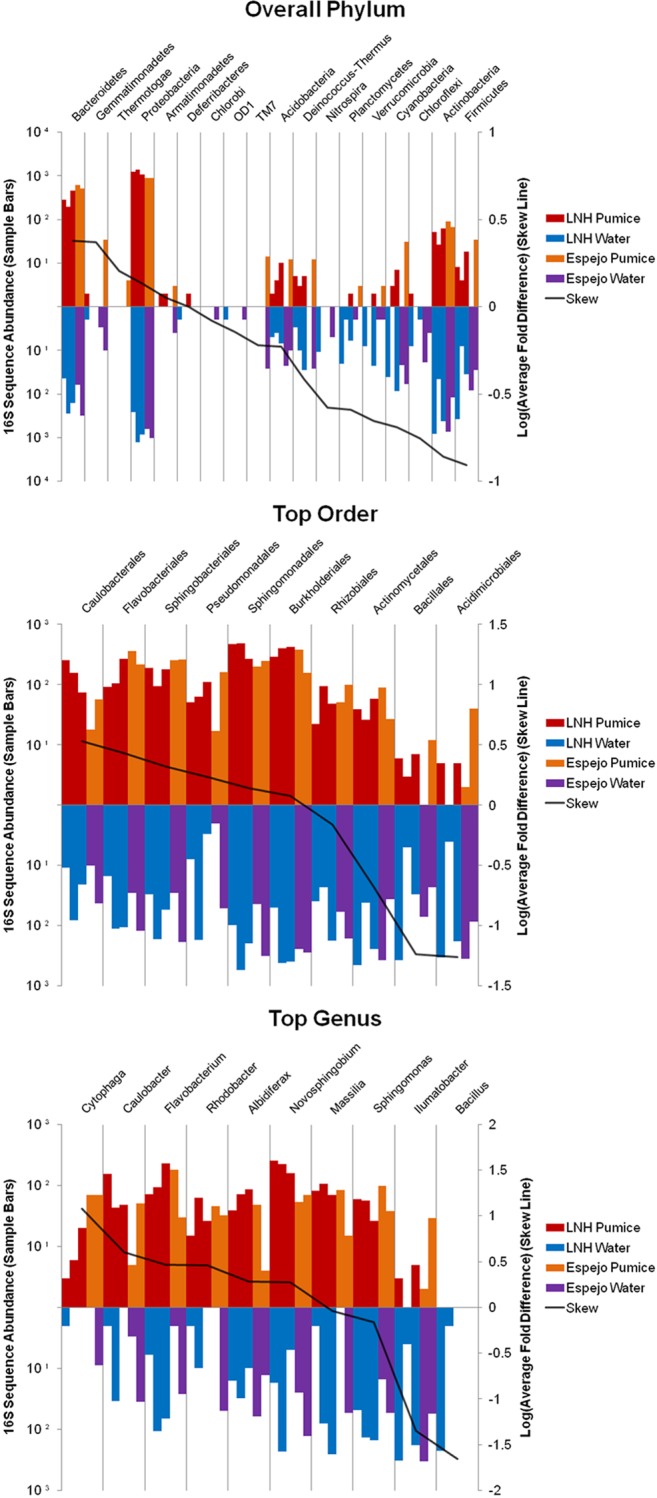
Taxa that were differentially represented in the pumice relative to the water (left side) and in the water relative to the pumice (right side) at the phylogenetic levels of phylum, order and genus. Overall there were eight bacterial orders found only on the pumice and not in the water, while there were 10 orders found in the water that were not found on the pumice. Overall there were 69 genera found only on pumice, while 100 genera were detected in the water but not on pumice (note that only the top 10 taxa that contributed most to distinguishing pumice from water column communities are depicted). The number on each bar indicates the total number of reads across all samples for each taxonomic group. Color coding indicates the two lakes that were sampled (Lake Nahuel Huapi [LNH] or Lake Espejo), while position order indicates the different sampling dates for each lake (three dates for LNH and two for Lake Espejo).
We next sought to determine if fresh pumice and colonized pumice had significant biogeochemical impacts on element cycling, with a particular focus on nutrient availability and dynamics in lake surface waters following the eruption. To do this, we first studied nutrient fluxes from fresh, free-falling pumice collected during the night of the initial eruption, allowing us to estimate the impact of the initial pumice input on availability of key nutrients. This fresh pumice released P at initial rates of 1.2 × 10−2 μmol P g−1 pumice day−1, but no release of N was detected, consistent with the pumice's observed P content of 0.009% (by mass) and its negligible N content. To quantify potential nutrient uptake by colonized pumice, we evaluated uptake kinetics using colonized pumice collected from Lake Espejo on 15 February 2012 (see Table S2 in the supplemental material). These kinetics results were combined with measurements of ambient nutrient concentrations in the Lake Espejo upper water column to estimate in situ nutrient fluxes per unit mass of pumice. We then extrapolated this value using the estimated midsummer abundance of floating pumice (see Materials and Methods). These calculations indicated that pumice in Lake Espejo was capable of taking up 1.92 to 19.2 μmol P m−2 day−1 and 33.3 to 333 μmol N m−2 day−1 (combined NH4 and NO3) (the range indicates the range of assumptions for midsummer pumice density). We then compared this pumice-associated microbial N and P uptake to that arising from microplankton in the surface waters of Lake Espejo (see Materials and Methods); these estimates indicated pelagic nutrient demands of 24 μmol P m−2 day−1 and 213 μmol N m−2 day−1. Thus, by midsummer, floating pumice communities were capable of generating substantial demands for P and N, equivalent to ∼8 to 82% and ∼15 to 156% of epilimnetic P and N demand, respectively.
DISCUSSION
Our data, while limited in temporal and spatial extents, indicate that microbial colonization of the novel substrata of floating pumice results in distinct assemblages that diverge significantly from ambient water column communities and that achieve a degree of structural similarity across habitats and times (Fig. 2), likely reflecting the stringent environmental conditions imposed by this dynamic habitat. This is particularly notable in what can be interpreted as convergent structures of the pumice communities for pairs of samples obtained from different lakes at different times (e.g., Lake Espejo on 20 September 2011 with Lake Nahuel Huapi on 16 January 2012 and Lake Nahuel Huapi on 1 December 2012 with Lake Espejo on 17 January 2012). This is especially noteworthy in that this similarity is observed despite the major seasonal differences encompassed by the sample dates: the surface water temperature in Lake Nahuel Huapi was ∼7.6°C on 9 September 2011 but 16.5°C on 16 January 2012. While more intensive investigations of future eruptions with greater replication would strengthen potential insights, these patterns suggest that the ecological challenges of proliferating on pumice are sufficiently stringent to override those imposed by seasonal change, resulting in a degree of convergence of community structure on pumice across seasons and lakes. The challenges appeared to be especially acute for photoautotrophs, given the underrepresentation of both cyanobacteria (Fig. 4) and eukaryotic photoautotrophs on pumice relative to the water column. This may reflect the highly erratic light regimes that such cells likely experience. When winds are calm, the pumice is dispersed and surfaces receive extremely high levels of irradiance, including high levels of UV radiation. However, sustained winds accumulate the pumice into thick rafts against the shoreline, and much of the pumice likely receives very low levels of incident radiation, perhaps insufficient to support photosynthesis. Assessments of such impacts can form the focus of future work on pumice-associated microbes.
The community structure analyses encompass a number of notable differences among lower-level taxa in occurrence and proliferation on pumice relative to the water. For example, the observed shift toward increased representation of the phylum Bacteroidetes in the pumice samples (Fig. 4; Table 1) is consistent with the known association of this group with suspended particles (24) due to an ability to adhere to surfaces (25), although in marine systems the colonized particles are of organic origin (“marine snow”). Both Cytophaga and Flavobacterium were disproportionately favored on pumice relative to the water (Fig. 4). Members of these taxa are well known for their ability to degrade high-molecular-weight organic compounds and are often associated with suspended detritus (26); these traits may allow members of this group to succeed in attached form on pumice particles, metabolizing extant refractory organic matter in the absence of abundant photoautotrophs in the pumice community. Their relatively high growth rates and ability to move across moist surfaces through gliding motility may also provide them an advantage as primary colonizers of new habitats (27), such as freshly deposited pumice. Similarly, the typical oligotrophic freshwater bacterium, Caulobacter, appeared to be favored on pumice. Caulobacter is known to have, in its complex cell cycle, a surface-associated stage that can be induced by light (28). Caulobacter is also an obligate aerobe, and growing on buoyant pumice ensures sufficient oxygen availability. The proliferation of Albidiferax ferrireducens (Proteobacteria) on pumice indicates that the pumice creates unique microenvironments in the pelagic zone, as this species is known to metabolize using iron as an electron acceptor (29), suggesting that, relative to the water column, the floating pumice provides a biogeochemically heterogeneous and distinctive habitat in which reducing conditions are present despite overall high levels of oxygenation of epilimnetic waters.
While limited replication and consideration of spatial and temporal variations prevent a definitive conclusion from being drawn, our measurements of pumice-associated nutrient fluxes do indicate a dynamic situation in which, as time progressed during the ensuing austral summer, microbial colonization shifted pumice from its initial impact as a source of nutrients due to leaching of P to a sink of nutrients due to biological uptake. This impact of pumice-associated nutrient uptake was observed even though half-saturation constants for the pumice were much higher than those generally observed for phytoplankton and bacterioplankton from oligotrophic lakes and oceans (30, 31), likely reflecting slow transport of nutrients across boundary layers of the large pumice particles. However, our estimates involve considerable uncertainty, especially with respect to estimates of areal abundance of pumice in the lakes. These were based on local reports (9) and subject to regional variation as well as unknown impacts of wind, topography, and watershed configuration that may have influenced the amount of pumice reaching each lake. Furthermore, the amount of pumice likely varied seasonally, as pumice became stranded on shore as lake levels decreased during summer. Likewise, pumice buoyancy has a limited lifetime, and thus unknown amounts of pumice sunk to lake depths during the ensuing summer. Nevertheless, we note that rafts of floating pumice can still be seen in these lakes, although at greatly reduced abundances relative to those in our study period immediately after eruption.
Overall, our data demonstrate the ability of diverse and distinct microbial taxa to establish and proliferate on floating pumice at biogeochemically significant levels, pointing to the biological suitability of this unusual habitat despite the seeming harshness of its irradiance and disturbance regimes. These findings support a recent contention that floating pumice may have served as an initial cradle for the emergence of life on early Earth (14).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the U.S. National Science Foundation, the NASA Astrobiology Institute, the Fondo Para la Investigación Científica y Tecnológica (Argentina FONCyT PICT2240, PICT1168), the CONICET-NSF Cooperation Program, and the National Geographic Society (NGS9001/11). J.J.E. acknowledges support from the Fulbright Foundation.
G. Olmedo and J. Raymond provided helpful comments on the manuscript. We also thank A. Glukhova for laboratory support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03160-14.
REFERENCES
- 1.Lindenmayer DB, Likens GE, Franklin JF. 2010. Rapid responses to facilitate ecological discoveries from major disturbances. Front Ecol Environ 8:527–532. doi: 10.1890/090184. [DOI] [Google Scholar]
- 2.Larson DW. 2011. Science after the volcano blew: research near Mount St. Helens proceeded despite bureaucratic hurdles, limited funding and an extremely hazardous environment. Am Sci 98:324–333. doi: 10.1511/2010.85.324. [DOI] [Google Scholar]
- 3.Campbell J, Bibb WF, Lambert MA, Eng S, Steigerwalt AG, Allard J, Moss CW, Brenner DJ. 1984. Legionella sainthelensi: a new species of Legionella isolated from water near Mount St. Helens. Appl Environ Microbiol 47:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balseiro EG, Modenutti BE, Queimalinos CP. 1997. Nutrient recycling and shifts in N:P ratio by different zooplankton structures in a South Andes lake. J Plankton Res 19:805–817. doi: 10.1093/plankt/19.7.805. [DOI] [Google Scholar]
- 5.Balseiro E, Modenutti B, Queimalinos C, Reissig M. 2007. Daphnia distribution in Andean Patagonian lakes: effect of low food quality and fish predation. Aquat Ecol 41:599–609. doi: 10.1007/s10452-007-9113-3. [DOI] [Google Scholar]
- 6.Corno G, Modenutti BE, Callieri C, Balseiro EG, Bertoni R, Caravati E. 2009. Bacterial diversity and morphology in deep ultraoligotrophic Andean lakes: role of UVR on vertical distribution. Limnol Oceanogr 54:1098–1112. doi: 10.4319/lo.2009.54.4.1098. [DOI] [Google Scholar]
- 7.Modenutti B, Balseiro E, Callieri C, Queimalinos C, Bertoni R. 2004. Increase in photosynthetic efficiency as a strategy of planktonic organisms exploiting deep lake layers. Freshw Biol 49:160–169. doi: 10.1046/j.1365-2427.2003.01169.x. [DOI] [Google Scholar]
- 8.Modenutti BE, Balseiro EG, Callieri C, Bertoni R, Queimalinos CP. 2005. Effect of UV-B and different PAR intensities on the primary production of the mixotrophic planktonic ciliate Stentor araucanus. Limnol Oceanogr 50:864–871. doi: 10.4319/lo.2005.50.3.0864. [DOI] [Google Scholar]
- 9.Gaitan JJ, Ayesa JA, Umaña F, Raffo F, Bran DB. 2011. Cartografía del área afectada por cenizas volcánicas en las provincias de Río Negro y Neuquén. INTA, S.C. de Bariloche, Argentina. [Google Scholar]
- 10.Modenutti BE, Balseiro EG, Elser JJ, Bastidas Navarro M, Cuassolo F, Laspoumaderes C, Souza MS, Diaz Villanueva V. 2013. Effect of volcanic eruption on nutrients, light, and phytoplankton in oligotrophic lakes. Limnol Oceanogr 58:1165–1175. doi: 10.4319/lo.2013.58.4.1165. [DOI] [Google Scholar]
- 11.Thiel M, Gutow L. 2005. The ecology of rafting in the marine environment. I. The floating substrata. Oceanogr Mar Biol Annu Rev 42:181–263. [Google Scholar]
- 12.Thiel M, Gutow L. 2005. The ecology of rafting in the marine environment. II. The rafting organisms and community. Oceanogr Mar Biol Annu Rev 43:279–418. [Google Scholar]
- 13.Ebbesmeyer CC, Ingraham WJ. 1999. Pumice and mines afloat on the sea. Oceanography 12:17–21. [Google Scholar]
- 14.Brasier MD, Matthewman R, McMahon S, Wacey D. 2011. Pumice as a remarkable substrate for the origin of life. Astrobiology 11:725–735. doi: 10.1089/ast.2010.0546. [DOI] [PubMed] [Google Scholar]
- 15.Iriondo M. 1989. Quaternary lakes of Argentina. Palaeogeogr Palaeoclimatol Palaeoecol 70:81–88. doi: 10.1016/0031-0182(89)90081-3. [DOI] [Google Scholar]
- 16.Wilson T, Stewart G, Bickerton H, Baxter T, Outes V, Villarosa G, Rovere E. 2013. Impacts of the June 2011 Puyehue-Cordon Caulle volcanic complex eruption on urban infrastructure, agriculture, and public health. GNS Sci Rep 2012 20:88. [Google Scholar]
- 17.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Austral Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.APHA. 2005. Standard methods for the examination of water and wastewater, 21st ed. American Water Works Association, Washington, DC. [Google Scholar]
- 23.Caravati E, Callieri C, Modenutti B, Corno G, Balseiro E, Bertoni R, Michaud L. 2010. Picocyanobacterial assemblages in ultraoligotrophic Andean lakes reveal high regional microdiversity. J Plankton Res 32:357–366. doi: 10.1093/plankt/fbp126. [DOI] [Google Scholar]
- 24.Rath J, Wu KY, Herndl GJ, DeLong EF. 1998. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol 14:261–269. doi: 10.3354/ame014261. [DOI] [Google Scholar]
- 25.Bauer M, Kube M, Teeling H, Richter M, Lombardot T, Allers E, Würdemann CA, Quast C, Kuhl H, Knaust F, Woebken D, Bischof K, Mussmann M, Choudhuri JV, Meyer F, Reinhardt R, Amann RI, Glöckner FO. 2006. Whole genome analysis of the marine Bacteroidetes “Gramella forsetii” reveals adaptations to degradation of polymeric organic matter. Environ Microbiol 8:2201–2213. doi: 10.1111/j.1462-2920.2006.01152.x. [DOI] [PubMed] [Google Scholar]
- 26.Kirchman DL. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 27.Fernández-Gómez B, Richter M, Schüler M, Pinhassi J, Acinas SG, González JM, Pedrós-Alió C. 2013. Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J 7:1026–1037. doi: 10.1038/ismej.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. 2007. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci U S A 104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu S, Chourey K, Reiche M, Nietzsche S, Shah MB, Neu TR, Hettich RL, Küsel K. 2013. Insights into the structure and metabolic function of microbes that shape pelagic iron-rich aggregates (“iron snow”). Appl Environ Microbiol 79:4272–4281. doi: 10.1128/AEM.00467-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith REH, Kalff J. 1982. Size dependent phosphorus uptake kinetics and cell quota in phytoplankton. J Phycol 18:275–284. doi: 10.1111/j.0022-3646.1982.00275.x,10.1111/j.1529-8817.1982.tb03184.x. [DOI] [Google Scholar]
- 31.Eppley RW, Rogers JN, McCarthy JJ. 1969. Half-saturation constants for uptake of nitrate and ammonium by marine phytoplankton. Limnol Oceanogr 14:912–920. doi: 10.4319/lo.1969.14.6.0912. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.