Abstract
To determine whether IDH1 mutations are present in primary and relapsed (local and distal) conventional central chondrosarcomas; and secondly, to assess if loss of p16/CDKN2A is associated with tumour grade progression, 102 tumour samples from 37 patients, including material from presenting and relapse events, were assessed. All wild-type cases for IDH1 R132 substitutions were also tested for IDH2 R172 and R140 alterations. The primary tumour and the most recent relapse sample were tested for p16/CDKN2A by interphase fluorescence in situ hybridisation. An additional 120 central cartilaginous tumours from different patients were also tested for p16/CDKN2A copy number. The study shows that if an IDH1 mutation were detected in a primary central chondrosarcoma, it is always detected at the time of presentation, and the same mutation is detected in local recurrences and metastatic events. We show that p16/CDKN2A copy number variation occurs subsequent to the IDH1 mutation, and confirm that p16/CDKN2A copy number variation occurs in 75 % of high grade central chondrosarcomas, and not in low grade cartilaginous tumours. Finally, p16/CDKN2A copy number variation is seen in both the IDH1 wild-type and mutant cartilaginous central tumours.
Keywords: Chondrosarcomas, IDH1, p16/CDKN2A, Sarcoma
Introduction
Isocitrate dehydrogenase 1 (IDH1), and IDH2 mutations were originally reported as frequent events in gliomas and acute myeloid leukaemia. We subsequently revealed the presence of the same mutations in at least 56 % of central chondrosarcomas: the mutations are present in all grades and the dedifferentiated variant but not in the other subtypes (peripheral and clear cell chondrosarcoma), or in soft tissue tumours [1, 6]. The mutations are also present in enchondromas, the benign central cartilaginous tumour considered to represent the precursor of central chondrosarcoma, and occur as early post-zygotic events in the common forms of multiple enchondromas, namely Ollier disease and Maffucci syndrome [3, 9]. These findings make a case for IDH1/2 mutations occurring early in the development of the non-syndromic disease and that the mutations represent driver mutations, to which the tumours are addicted for survival. Nevertheless, to provide further evidence for this, we now evaluate if the mutations always occur in the presenting tumour, and if they persist in recurrent and metastatic disease. This is an important question if drugs, which may be developed against the mutant protein, are to be effective [10].
Disease progression in IDH1/2 mutant-bearing tumours is likely to depend on additional contributing genetic events. Loss of copy number of p16/CDKN2A is a well recognised tumour suppressor gene involved in cancer oncogenic process, and has been reported to be associated with disease progression of cartilaginous tumours. Specifically, p16/CDKN2A has been found to occur in high grade chondrosarcomas but rarely in chondrosarcoma grade (G) I and not in enchondromas [5, 13]. Furthermore, we subsequently revealed that homozygous p16/CDKN2A deletions were only detected in high grade chondrosarcomas (GII, GIII and dedifferentiated) but not in GI central tumours, and that these genetics alterations were detected in both IDH1 mutant and wild-type (WT) central chondrosarcomas [12]. Therefore, we wished to determine if loss of p16/CDKN2A in relapse disease correlates with progression to a higher grade of tumour.
Material and methods
One hundred and two samples from 37 patients, with the associated demographic data, and the interval between the primary tumour and first relapse event, were retrieved from the files of the Royal National Orthopaedic Hospital NHS Trust. Haematoxylin and eosin-stained sections from all selected tumours were reviewed (MFA, AMF, RT). Age and gender of the 37 patients with their tumour grade are shown in Table 1.
Table 1.
Characteristics of 37 patients with multiple samples
| Case | Age | Gender | Site primary | Number of samples | Grade primary | Grade relapses | IDH1 status | CDKN2A/p16 | Interval from primary to 1st relapse (months) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 32 | female | sacrum | 3 | G2 | G2 | WT | loss > further loss | 31 |
| 2 | 80 | male | femur | 4 | Dediff | Dediff | R132H | normal > loss | 24 |
| 3 | 35 | male | pelvis | 2 | G2 | G2 | WT | loss | 51 |
| 4 | 50 | male | vertebra | 3 | G2 | G2 | WT | NI | 102 |
| 5 | 69 | female | rib | 4 | G2/3 | G2/3 | WT | NI | 15 |
| 6 | 41 | female | sternum | 2 | G2 | G3 | WT | normal | 101 |
| 7 | 39 | female | tibia | 2 | G2 | G2 | WT | normal | 88 |
| 8 | 57 | male | rib | 4 | G2 | G2 | R132C | normal | 8 |
| 9 | 30 | male | scapula | 2 | G2 | Dediff | WT | loss > polysomic | 17 |
| 10 | 41 | male | vertebra | 3 | G3 | G3 | WT | loss | 27 |
| 11 | 44 | male | pelvis | 3 | G3 | G3 | R132G | polysomic | 19 |
| 12 | 22 | male | sacrum | 3 | G2 | G2 | WT | loss | 12 |
| 13 | 51 | male | humerus | 3 | G3 | G3 | WT | normal > polysomic | 9 |
| 14 | 74 | male | femur | 2 | G2 | G2 | R132G | loss | 38 |
| 15 | 42 | female | hand | 2 | G2 | G2 | R132S | NI | 67 |
| 16 | 39 | male | humerus | 3 | G2 | G3 | R132C | normal > loss | 15 |
| 17 | 43 | male | femur | 2 | G2 | G2 | R132C | normal | 57 |
| 18 | 64 | female | tibia | 3 | G1 | G1 | WT | NI | 14 |
| 19 | 55 | male | hand | 6 | G2 | G3 | R132G | normal | 15 |
| 20 | 28 | female | pelvis | 2 | G2 | G2 | WT | loss | 34 |
| 21 | 31 | male | scapula | 5 | G2 | G3 | WT | normal > loss | 15 |
| 22 | 69 | female | femur | 3 | G2 | G3 | R132G | NI | 13 |
| 23 | 43 | male | vertebra | 4 | G2 | G2 | WT | normal | 11 |
| 24 | 34 | male | humerus | 2 | G2 | G3 | WT | normal > loss | 16 |
| 25 | 50 | male | humerus | 3 | G3 | G3 | WT | loss | 12 |
| 26 | 77 | male | tibia | 2 | G3 | G3 | R132L | loss | 33 |
| 27 | 72 | male | femur | 2 | Dediff | Dediff | R132H | loss | 6 |
| 28 | 58 | female | sacrum | 2 | G2 | G2 | WT | normal | 27 |
| 29 | 36 | female | humerus | 2 | G1 | G1 | R132C | normal | 15 |
| 30 | 51 | male | femur | 3 | Dediff | Dediff | R132H | loss | 5 |
| 31 | 52 | male | pelvis | 2 | Dediff | Dediff | R132C | polysomic | 12 |
| 32 | 55 | male | femur | 2 | Dediff | Dediff | R132G | loss | 4 |
| 33 | 30 | female | tibia | 3 | G2 | G2 | WT | normal | 48 |
| 34 | 68 | male | femur | 2 | Dediff | Dediff | R132S | polysomic | 1 |
| 35 | 61 | male | femur | 2 | G2 | G2 | R132H | normal > loss | 23 |
| 36 | 65 | male | rib | 2 | G2 | Dediff | WT | NI | 26 |
| 37 | 29 | female | femur | 3 | G2 | G2 | R132C | normal | 127 |
G grade, Dediff dedifferentiated chondrosarcomas, WT wild-type
An additional 120 central conventional cartilaginous tumours from our previous IDH study were analysed for p16/CDKN2A copy number [1]. This cohort included 61 low grade cartilaginous tumours (enchondromas and chondrosarcomas GI), 2 chondrosarcomas GI with transition to GII, 30 chondrosarcomas GII, 6 chondrosarcomas GIII and 21 dedifferentiated chondrosarcomas.
Genomic DNA was extracted from material that was at least 60 % tumour-rich and was analysed using PCR amplification followed by capillary sequencing and/or realtime PCR as previously described [1]. Sections from the primary tumour and the most recent recurrence were analysed by fluorescence in situ hybridisation (FISH) for p16/CDKN2A (9p21) using commercially available probes (Vysis, Abbott Molecular, Illinois USA) as previously described [2]. Loss of p16/CDKN2A was defined as: (i) monosomy (1 p16/CDKN2A and 1 centromeric signal); (ii) heterozygous deletion (loss of 1 copy of p16/CDKN2A in the presence of 2 centromeric signals) and (iii) homozygous deletion (loss of 2 copies of p16/CDKN2A in the presence of 1 or 2 centromeric signals) in at least 15 % of the nuclei analysed (minimum count of 50 consecutive non-overlapping nuclei). Polysomy was defined as more than 2 copies of p16/CDKN2A/CEP 9 in at least 15 % of the nuclei analysed.
Results
One hundred and two tumour samples from 37 patients were analysed for IDH1 R132 of which 48 (49 %) were found to harbour a R132 IDH1 mutation. Thirty of these 48 tumours represented disease from relapse events (27 local recurrences and 3 metastatic events). An IDH1 mutation was detected in the presenting and relapse samples from each of these 18 patients. All samples from a given patient in this group had the same IDH1 alteration (Fig. 1a.). If a tumour at presentation was found to be wild-type (WT) for IDH1 R132 mutations, the relapse samples were also WT (19/37 patients). In IDH1 WT cases, at least 1 sample, from each of these patients, was also screened for both IDH2 R172 and R140 mutations. No IDH2 (R172 or R140) mutations were detected in this cohort.
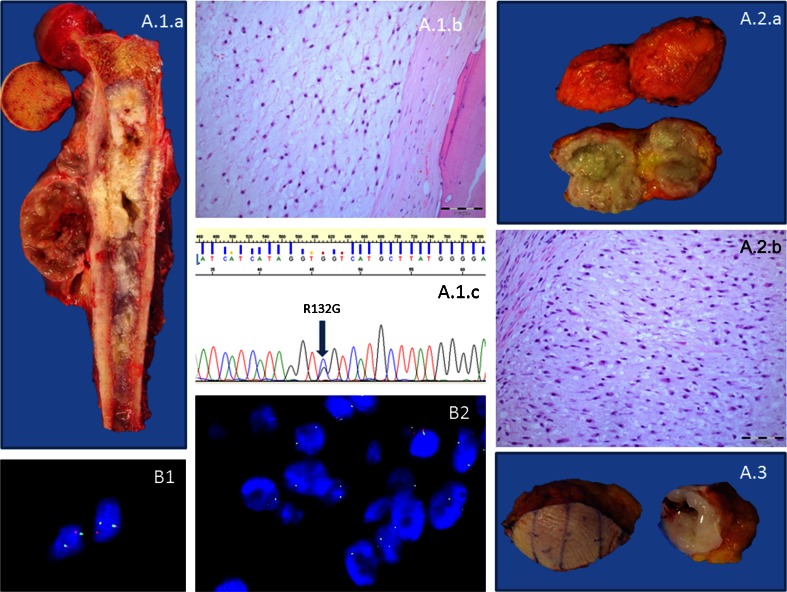
Fig. 1.
a Case 22 showing the primary (A.1.a) excision of a chondrosarcoma GII, (A.1.b) harbouring a R132G substitution (A.1.c) and 2 subsequent relapse events in the chest wall (A.2.a, A.2.b and A.3). b Case 30. Diploid p16/CDKN2A copy number (B1) and heterozygous loss of p16/CDKN2A (B2)
Details of the interval between the primary treatment (curettage or wide excision) of the presenting tumour and the first recurrence is given in Table 1. The number of relapse events per patient ranged from 1 to 5 (average 1.9).
Analysis of the p16/CDKN2A gene locus by FISH gave informative results from both the primary tumour and the most recent relapse in 31 of the 37 patients. In contrast to the persistent presence of IDH1 mutations throughout tumour progression, p16/CDKN2A copy number variation only occurred in high grade chondrosarcomas (GII, GIII or dedifferentiated neoplasms). Of the 21 tumour sets (from 31 informative cases) in which p16/CDKN2A copy number variation was identified in either the primary and/or relapse event, 11 harboured an IDH1 mutation and 10 were WT for IDH1 and IDH2, indicating that change in p16/CDKN2A copy number is equally common in IDH1 mutant and WT central conventional cartilaginous tumours.
Of the 31 informative tumour sets, 23 revealed the same p16/CDKN2A copy number in both samples, and 8 had discordant copy number (vide infra). The former included 10 with normal copy number of p16/CDKN2A, 10 with loss of p16/CDKN2A, and 3 with polysomy for chromosome 9. In only 2 of these tumour sets did the grade of the tumour progress; in both cases from GII to GIII, both with normal copy number of p16/CDKN2A (Table 1). There were only 2 chondrosarcoma GI which recurred, 1 was non-informative for FISH, and the other revealed a normal copy number of p16/CDKN2A. Within this group, the samples from patient 30 were particularly noteworthy in that, the presenting tumour was a dedifferentiated chondrosarcoma in which the normal (diploid) p16/CDKN2A copy number was detected in the well-differentiated component (Fig. 1. b1; Table 1) and a homozygous deletion was detected in both the dedifferentiated component of the presenting and the metastatic tumour (Fig. 1. b2).
Of the 8 sample sets from the 31 patients which revealed discordant p16/CDKN2A copy number, loss of p16/CDKN2A was observed in the relapse event and not in the primary tumour in 5 cases (3 of which also harboured IDH1 mutation). In tumours from 2 patients, polyploidy of chromosome 9 was observed in the relapse event whereas in the primary tumour, the normal copy number was seen in 1 case and monosomy in the other. In the remaining case, loss of p16/CDKN2A converted from being heterozygous in the primary tumour to being homozygous in the relapse sample. In 4/8 (50 %) of these patients, progression of the chondrosarcoma grade occurred in the relapse. In 3, the primary tumour showed normal copy number, and progression to a higher morphological grade (GII to GIII) was accompanied by p16/CDKN2A loss. The fourth tumour which presented as a chondrosarcoma GII was monosomic for chromosome 9 and progressed to a dedifferentiated tumour, in which the tumour cells were polysomic for chromosome 9.
The additional 120 central conventional cartilaginous tumours from 120 different patients were analysed for p16/CDKN2A by FISH. Of the 61 low grade tumours, all revealed normal diploid p16/CDKN2A copy number. Loss of p16/CDKN2A was detected in 25 % (8/30) of the CHS GII, including 2 cases diagnosed as GI with transition to GII, and 50 % of the CHS GIII (3/6). The 62 % of the dedifferentiated chondrosarcomas (13/21) also revealed p16/CDKN2A loss. Polysomy of p16/CDKN2A was detected in 2/30 GII, 1/6 GIII and 6/21 dedifferentiated chondrosarcomas.
Further analysis of 3/21 dedifferentiated chondrosarcomas where material was available/suitable for analysis, there was normal diploid p16/CDKN2A copy number in the well-differentiated component and p16/CDKN2A loss in the dedifferentiated component.
Discussion
In this study, we confirm the data published in our previous report showing that IDH1 mutations occur in ∼50 % of solitary conventional central chondrosarcomas, that the IDH1 mutations are found in all grades of this subtype of chondrosarcoma, that the R132C mutation is the most common genetic alteration in this gene, that IDH2 mutations are significantly less common than IDH1 mutations and that to date an IDH2 R140 mutation, which is seen in acute myeloid leukaemia, is not found in cartilaginous tumours [1]. In addition, we now show that when an IDH1 mutation is present in a solitary primary central conventional chondrosarcoma, the same mutation is always detected in local recurrences and metastatic events. Furthermore, a mutation is never detected in a locally recurrent tumour or metastatic lesion when it is not detected in the presenting tumour. These data add weight to existing evidence that the R132 IDH1 mutations occur early in the genesis of cartilaginous tumours and are therefore important driver alterations. This provides a strong argument that the cartilaginous tumours harbouring the genetically altered IDH1 are good candidates for the selective inhibitors of the metabolic enzyme, IDH1 [10].
Although IDH1/IDH2 mutations appear to be an important genetic event in the pathogenesis of enchondromas and central chondrosarcomas, other mutations are likely to exert an impact on the biological behaviour of the disease. To date, two important tumour suppressor genes, p16/CDKN2A and TP53 have been implicated in the development of high grade chondrosarcoma [5, 11, 12]. Genetic alterations of both of these genes have been detected previously in chondrosarcomas [4, 5, 12]. In the case of TP53, inactivation has been shown to occur as a result of chromosomal loss and point mutations, and TP53 mutations have been detected in both IDH1 WT and mutant chondrosarcomas [8, 12]. Herein, we demonstrate that alteration in p16/CDKN2A copy number occurs exclusively in high grade central cartilaginous tumours, and that the percentage of cases with alteration of copy number of p16/CDKN2A increases with grade. Specifically, we show that 75 % (67 tumours from 89 patients) of central high grade chondrosarcomas reveal loss or gain (polysomy) of p16/CDKN2A in at least one sample (data generated from both cohorts). We also show that loss of p16/CDKN2A is found in both IDH1 WT and mutant cartilaginous central tumours, indicating that the two genetic events are unrelated.
We consider that our findings support the concept that alteration of p16/CDKN2A copy number occurs subsequent to the acquisition of the IDH1 mutation. This is supported by our findings that in 60 low grade central conventional tumours (enchondroma/low grade chondrosarcoma), obtained from patients with a single tumour sample, none had p16/CDKN2A copy number variation. This concurs with previously published data from smaller cohorts which report that loss of p16/CDKN2A is rarely present in enchondromas and low grade chondrosarcomas, whereas IDH1 and IDH2 mutations are found in tumours of all grades of chondrosarcomas and in benign conventional central cartilaginous tumours [12, 13]. Our findings indicate that if a tumour shows p16/CDKN2A copy number variation (gain or loss), it should be managed as a high grade chondrosarcoma. As grading of chondrosarcoma has been shown to lack reproducibility [7] assessment of alteration in copy number of p16/CDKN2A can be used to complement histological grading of chondrosarcoma. The presence of alteration in copy number argues strongly for a diagnosis of high grade disease, although the absence of such abnormality is not helpful as such changes are detected in ∼75 % of high grade chondrosarcoma. Future studies would be helpful to identify a biomarker to distinguish the remaining 25 % of high grade chondrosarcomas from low grade disease.
We conclude that IDH1 mutations represent a recurrent genetic event in solitary central chondrosarcomas, that they occur early in disease development and persist during tumour recurrence and metastatic disease thereby adding to the existing evidence that such mutations represent drivers of disease. The p16/CDKN2A copy number variation occurs in at least 50 % of IDH1 mutant-bearing high grade chondrosarcomas, and the findings support the concept that the p16/CDKN2A alterations are likely to be associated with disease progression. Assessment of p16/CDKN2A copy number can be used as an ancillary test to help reach a conclusion about the grade of a chondrosarcoma thereby providing additional information on which to decide clinical management of this disease.
Acknowledgments
This work was funded by the Pathological Society of Great Britain and Ireland. The material was obtained from the Stanmore Musculoskeletal Research Programme and Biobank. Support was provided by the National Institute for Health Research, University College London Hospitals Biomedical Research Centre and the UCL Experimental Cancer Centre, to AMF. Consenting of patients and data collection was performed by Ru Grinell at the RNOH NHS Trust. We are grateful to the patients for participating in the research and to the clinicians and support staff of the London Sarcoma Service involved in their care.
Conflict of interest
The authors have no financial or commercial interests that are relevant to this publication.
References
- 1.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O’Donnell P, Grigoriadis A, Diss T, Eskandarpour M, Presneau N, Hogendoorn PC, Futreal A, Tirabosco R, Flanagan AM. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 2.Amary MF, Berisha F, Bernardi Fdel C, Herbert A, James M, Reis-Filho JS, Fisher C, Nicholson AG, Tirabosco R, Diss TC, Flanagan AM. Detection of SS18-SSX fusion transcripts in formalin-fixed paraffin-embedded neoplasms: analysis of conventional RT-PCR, qRT-PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod Pathol. 2007;20:482–496. doi: 10.1038/modpathol.3800761. [DOI] [PubMed] [Google Scholar]
- 3.Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, McCarthy S, Fantin VR, Straley KS, Lobo S, Aston W, Green CL, Gale RE, Tirabosco R, Futreal A, Campbell P, Presneau N, Flanagan AM. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011;43:1262–1265. doi: 10.1038/ng.994. [DOI] [PubMed] [Google Scholar]
- 4.Antonelli M, Buttarelli FR, Arcella A, Nobusawa S, Donofrio V, Oghaki H, Giangaspero F Prognostic significance of histological grading, p53 status, YKL-40 expression, and IDH1 mutations in pediatric high-grade gliomas J Neurooncol. doi: 10.1007/s11060-010-0129-5 [DOI] [PubMed]
- 5.Asp J, Sangiorgi L, Inerot SE, Lindahl A, Molendini L, Benassi MS, Picci P. Changes of the p16 gene but not the p53 gene in human chondrosarcoma tissues. Int J Cancer. 2000;85:782–786. doi: 10.1002/(SICI)1097-0215(20000315)85:6<782::AID-IJC7>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 6.Damato S, Alorjani M, Bonar F, McCarthy SW, Cannon SR, O’Donnell P, Tirabosco R, Amary MF, Flanagan AM. IDH1 mutations are not found in cartilaginous tumours other than central and periosteal chondrosarcomas and enchondromas. Histopathology. 2012;60:363–365. doi: 10.1111/j.1365-2559.2011.04010.x. [DOI] [PubMed] [Google Scholar]
- 7.Eefting D, Schrage YM, Geirnaerdt MJ, Le Cessie S, Taminiau AH, Bovee JV, Hogendoorn PC. Assessment of interobserver variability and histologic parameters to improve reliability in classification and grading of central cartilaginous tumors. Am J Surg Pathol. 2009;33:50–57. doi: 10.1097/PAS.0b013e31817eec2b. [DOI] [PubMed] [Google Scholar]
- 8.Ho L, Stojanovski A, Whetstone H, Wei QX, Mau E, Wunder JS, Alman B. Gli2 and p53 cooperate to regulate IGFBP-3-mediated chondrocyte apoptosis in the progression from benign to malignant cartilage tumors. Cancer Cell. 2009;16:126–136. doi: 10.1016/j.ccr.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Pansuriya TC, Oosting J, Krenacs T, Taminiau AH, Verdegaal SH, Sangiorgi L, Sciot R, Hogendoorn PC, Szuhai K, Bovee JV. Genome-wide analysis of Ollier disease. Is it all in the genes? Orphanet J Rare Dis. 2011;6:2. doi: 10.1186/1750-1172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, Kunii K, Pedraza A, Schalm S, Silverman L, Miller A, Wang F, Yang H, Chen Y, Kernytsky A, Rosenblum MK, Liu W, Biller SA, Su SM, Brennan CW, Chan TA, Graeber TG, Yen KE, Mellinghoff IK. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrage YM, Lam S, Jochemsen AG, Cleton-Jansen AM, Taminiau AH, Hogendoorn PC, Bovee JV. Central chondrosarcoma progression is associated with pRb pathway alterations: CDK4 down-regulation and p16 overexpression inhibit cell growth in vitro. J Cell Mol Med. 2009;13:2843–2852. doi: 10.1111/j.1582-4934.2008.00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarpey PS, Behjati S, Cooke SL, Van Loo P, Wedge DC, Pillay N, Marshall J, O’Meara S, Davies H, Nik-Zainal S, Beare D, Butler A, Gamble J, Hardy C, Hinton J, Jia MM, Jayakumar A, Jones D, Latimer C, Maddison M, Martin S, McLaren S, Menzies A, Mudie L, Raine K, Teague JW, Tubio JM, Halai D, Tirabosco R, Amary F, Campbell PJ, Stratton MR, Flanagan AM, Futreal PA. Frequent mutation of the major cartilage collagen gene COL2A1 in chondrosarcoma. Nat Genet. 2013;45:923–926. doi: 10.1038/ng.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Beerendonk HM, Rozeman LB, Taminiau AH, Sciot R, Bovee JV, Cleton-Jansen AM, Hogendoorn PC. Molecular analysis of the INK4A/INK4A-ARF gene locus in conventional (central) chondrosarcomas and enchondromas: indication of an important gene for tumour progression. J Pathol. 2004;202:359–366. doi: 10.1002/path.1517. [DOI] [PubMed] [Google Scholar]