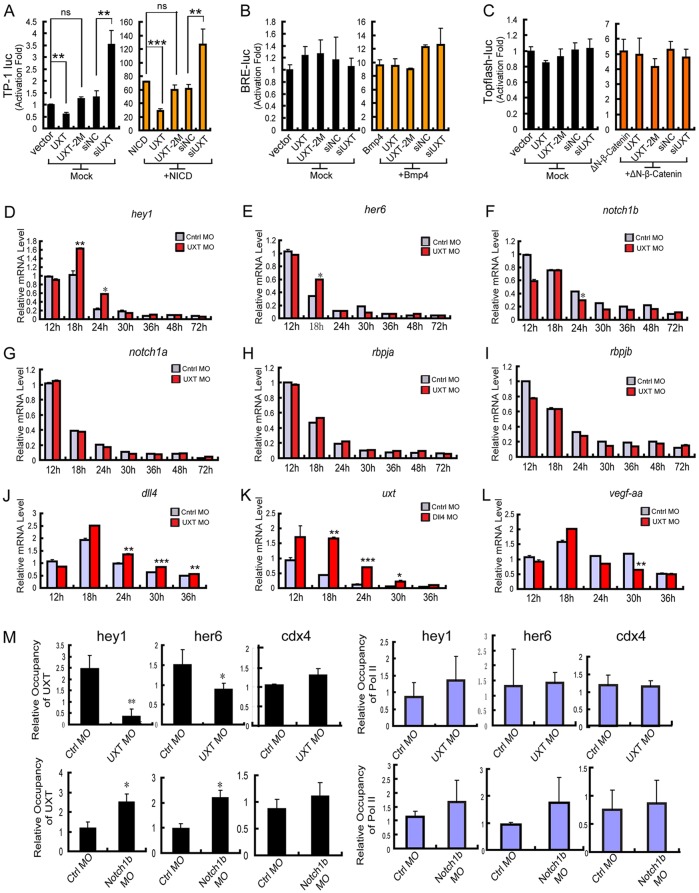
Fig. 5.
UXT attenuates the expression of the Notch target genes. (A) UXT impairs the TP-1 luciferase reporter of Notch signaling. The indicated plasmids and/or siRNAs were transfected into Cos-7 cells together with TP-1 reporter plasmids, in the presence or absence of NICD. (B) UXT does not influence BMP signaling. The indicated plasmids and/or siRNAs were transfected into Cos-7 cells together with the BRE reporter plasmids. After transfection, cells were starved for 12 h and then treated with or without BMP4 (10 ng). (C) UXT does not influence WNT signaling. The indicated plasmids and/or siRNAs were transfected into HEK293T cell lines together with the TOPflash reporter plasmids, in the presence or absence of ΔN-β-catenin. Data in A-C were normalized to empty vectors and are presented as the mean±s.e.m. (n=3). (D,E) UXT attenuates the expression of Notch target genes. The control and UXT-deficient zebrafish embryos (from 12 hpf to 72 hpf) were harvested, and the expression of hey1 (D) and her6 (E) were measured by RT-PCR. (F-J) UXT does not influence the expression of notch, rbpja or rbpjb but does affect dll4. The control and UXT-deficient zebrafish embryos (from 12 hpf to 72 hpf) were harvested, and the expression of notch1a (G), notch1b (F), rbpja (H), rbpjb (I) and dll4 (J) were measured by RT-PCR. (K) DLL4 attenuates the expression of UXT. The control and DLL4-deficient zebrafish embryos (from 12 hpf to 36 hpf) were harvested, and the expression of uxt was measured by RT-PCR. (L) The relative mRNA expression level of vegfaa in control and UXT-deficient embryos (from 12 hpf to 36 hpf). Data shown in D-L are the mean±s.e.m. (at least three independent experiments). (M) E-ChIP assays at 24 hpf. Embryos injected with 4 ng of control MO, 4 ng of UXT MO or Notch1b MO were probed with the antibody against UXT and polymerase II, respectively. The data were normalized on the basis of the corresponding input control and are presented as the mean±s.e.m. (at least three independent experiments). For all panels, *P<0.05; **P<0.01; ***P<0.001; ns, non-significant versus the corresponding control or as indicated.