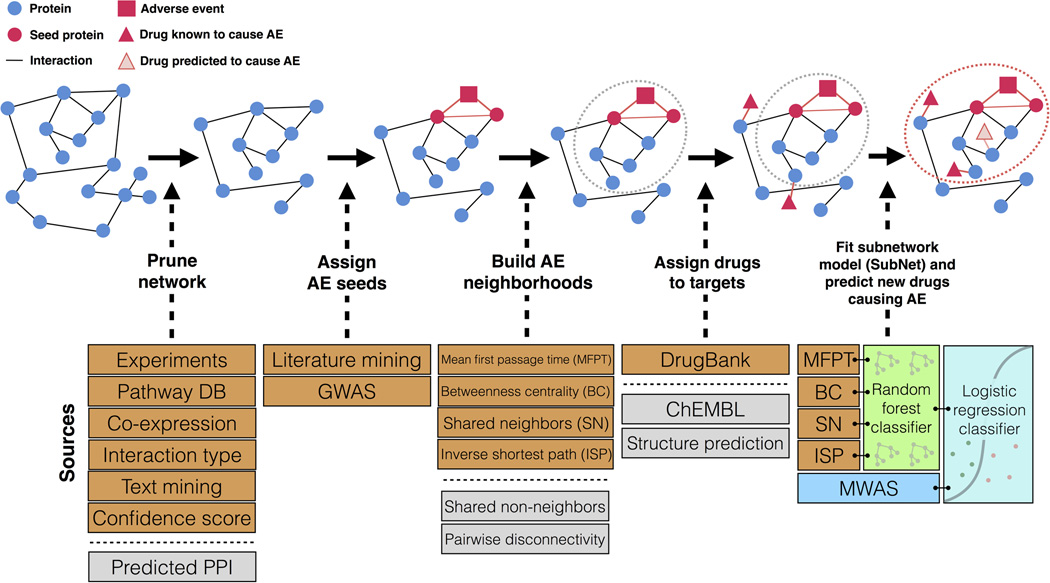
Figure 1.
Overview of Modular Assembly of Drug Safety Subnetworks (MADSS). Orange boxes indicate data sources used in this analysis. Gray boxes indicate additional data sources not used in this study but supported by the method. Beginning with a human protein-protein interaction network (interactome) built from such data as experimental evidence, metabolic pathway databases, text mining, and interactions predicted from co-expression data, we isolate all medium-confidence interactions and above. Seed proteins with demonstrated genetic links to the adverse event (AE) are subsequently annotated. We next apply four adapted network analysis functions to score all proteins in the interactome on their connectivity to the seed set. Proteins with high scores embody an AE neighborhood (gray dotted circle); drugs targeting proteins in this subnetwork are predicted to elicit AEs. We assign positive and negative control drugs to their highest-scoring target. We then combine the four AE neighborhoods (one for each pairwise network function) by training a random forest classifier to generate a subnetwork (SubNet) model (red dotted circle). We integrate MWAS and systems pharmacology (SubNet) models using a logistic regression classifier to predict drug safety.