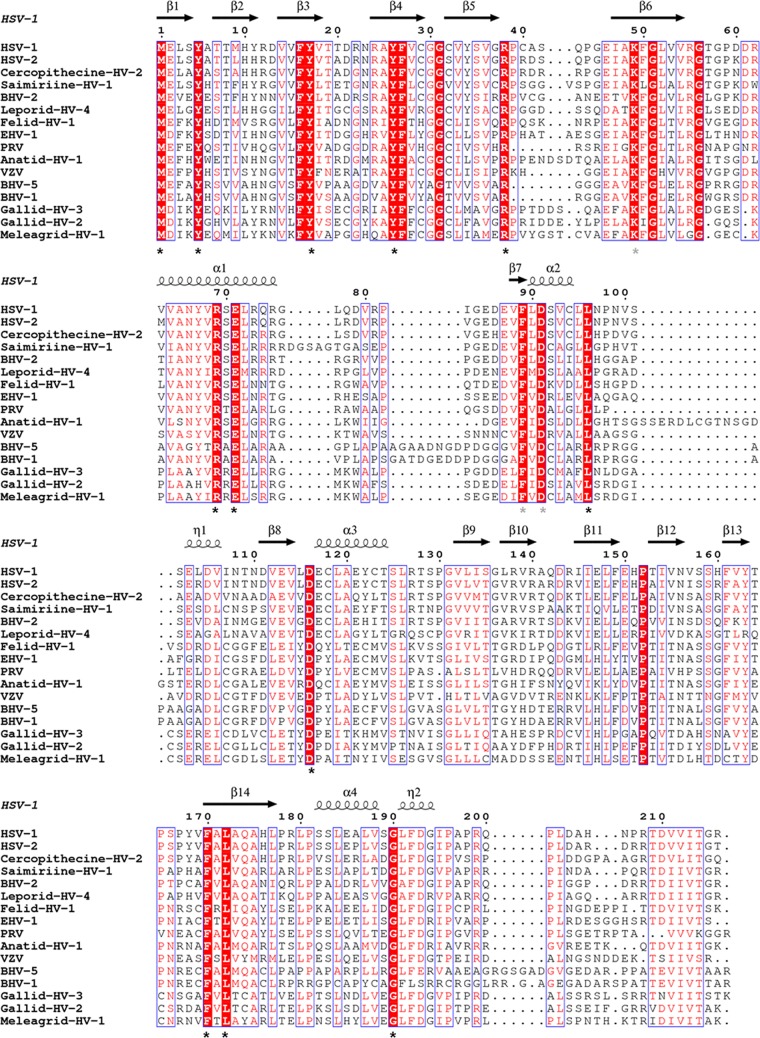
FIG 2.
Multiple-sequence alignment of UL21 homologs from 16 alphaherpesviruses. Sequence alignment was generated and analyzed using ClustalW (32) and ESPRIPT (33) using sequences from GenBank accession numbers ACM62243, AEV91359, AAU88086, ADO13807, AAK69349, AFR32463, ACT88337, AAT67298, AAA47475, ABO26208, AAT07796, AAR86141, CAA88112, AEI00223, AAF66756, and AAG45758. Only the alignment of residues corresponding to residues 1 to 216 of HSV UL21N is shown. The secondary structure of HSV-1 UL21 is shown above the aligned sequences. Similar residues are shown in red text. Identical residues are boxed in red with white text, and those exposed on the surface of UL21N are marked by asterisks. Gray asterisks identify conserved residues that are surface exposed in the model but are likely obscured in the protein by the unresolved loop containing residues 76 to 87.