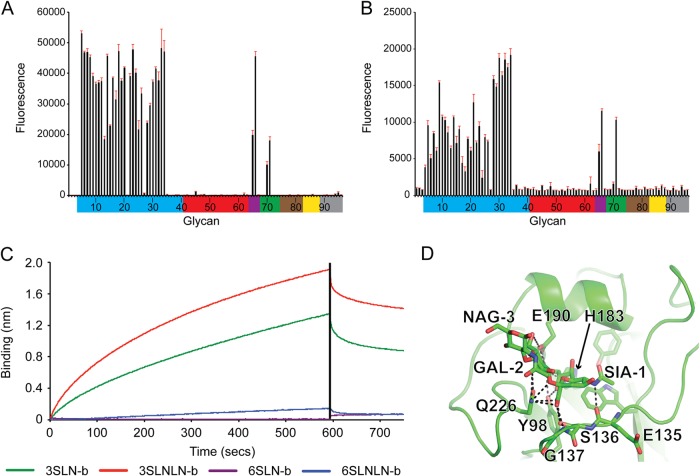
FIG 3.
Glycan binding to seal recHA. (A and B) Glycan array analysis of seal11 recHA (A) and A/harbor seal/New Hampshire/179629/2011 (B). Glycans on the microarray are grouped according to SA linkage: α2-3 SA (blue), α2-6 SA (red), α2-6–α2-3 mixed SA (purple), N-glycolyl SA (green), α2-8 SA (brown), β2-6 and 9-O-acetyl SA (yellow), and asialo glycans (gray). The error bars reflect the standard error in the signal for six independent replicates on the array. The structures of the numbered glycans are found in Table 2. Specific glycan structures that were used in biosensor assays are represented on the array as glycans 19/20, 22, 52/53, and 56. (C) The binding kinetics of seal11 recHA protein to specific biotinylated glycans, 3-SLN-b, 3-SLNLN-b, 6-SLN-b, and 6-SLNLN-b, immobilized on streptavidin-coated biosensors were analyzed by BLI. The black vertical line indicates the switch in the experiment from collecting association to collecting dissociation data. (D) 3-SLN glycan interactions with seal11 HA RBS. Seal HA is shown in cartoon form, while 3-SLN and interacting HA residues are shown as sticks. Hydrogen bonds between the glycan and HA are shown as dashed lines. The structural figure was generated with MacPyMol (83).