FIG 3.
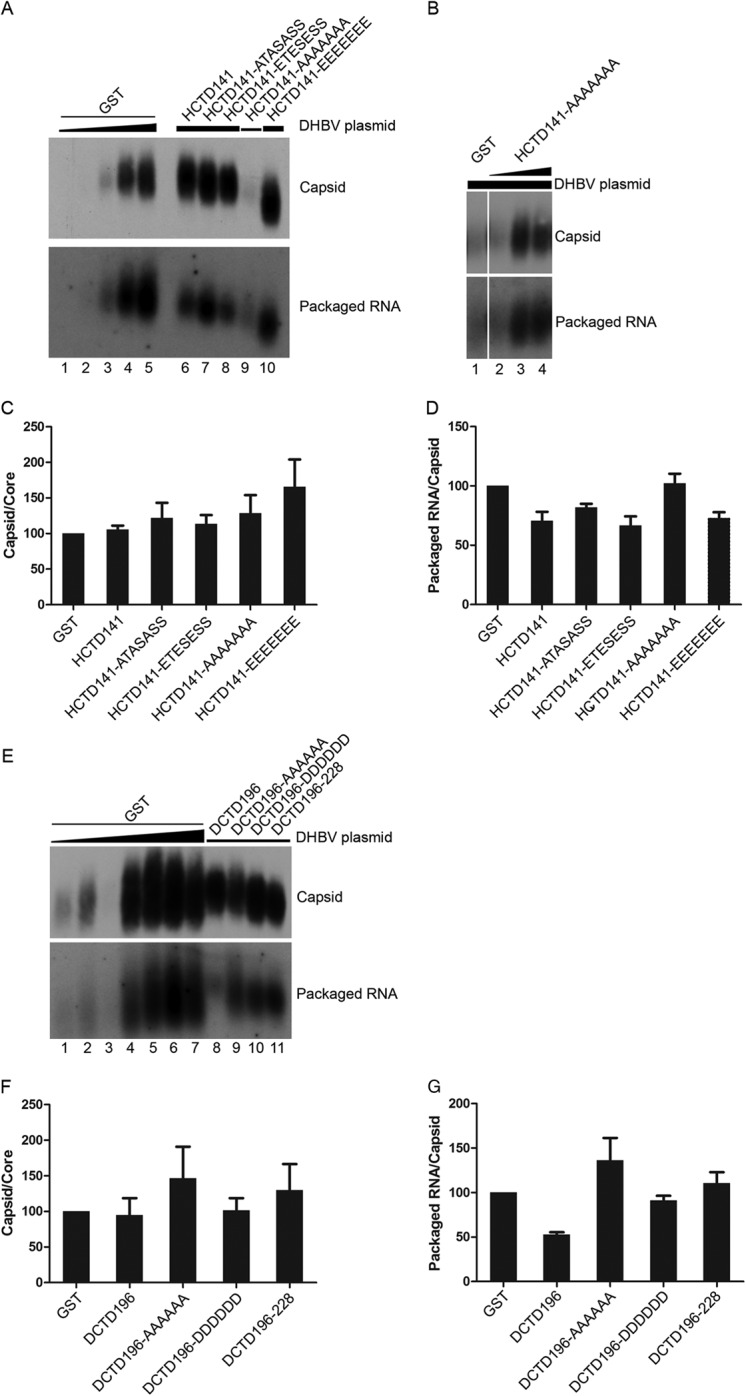
Effect of HCTD and DCTD on DHBV capsid assembly and pgRNA packaging. HEK293 cells were cotransfected as described in the legend to Fig. 2. Cytoplasmic capsids were analyzed by native agarose gel electrophoresis and transferred to a nitrocellulose membrane. A radiolabeled, plus-strand-specific DHBV riboprobe was used to detect the packaged pgRNA (A, B, and E, bottom), and the capsid protein on the same membrane (A, B, and E, top) was detected by an anti-DHBc antibody. Transfections shown in panels A and E were the same as those shown in Fig. 2A and D, respectively. Lanes 1 to 4 in panel B shows the same transfections as in lane 1 and lanes 11 to 13 in Fig. 2C. (C and F) The relative levels of assembled capsids from the GST-HCTD (A) or GST-DCTD (E) cotransfections were normalized to those of the total core protein detected by SDS-PAGE (shown in Fig. 2) (capsid/core) to estimate the assembly capability of DHBc. The capsid/core ratios from GST-HCTD (A) or GST-DCTD (E) were compared to that of the control (GST) transfection that is set to 100 (lane 4 in panel A and lane 2 in panel E). (D and G) The relative levels of packaged pgRNA were normalized to those of assembled capsids (packaged pgRNA/capsid) in the same manner. The packaged pgRNA/capsid ratios from the GST-HCTD (A) or GST-DCTD (E) cotransfections were compared to that of the control (GST) transfection that is set to 100 (lane 4 in panel A and lane 2 in panel E) to estimate DHBV RNA packaging capability. Data are presented as means and SEM from three independent experiments.
